8VAL
 
 | |
8VAQ
 
 | |
8VAP
 
 | |
8VAM
 
 | |
8VAS
 
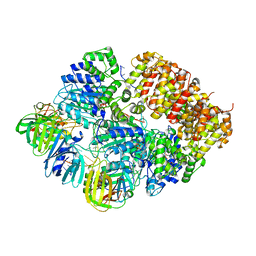 | |
7U1A
 
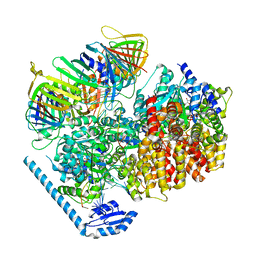 | | RFC:PCNA bound to dsDNA with a ssDNA gap of six nucleotides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA - Primer, DNA - Template, ... | | Authors: | Liu, X, Gaubitz, C, Pajak, J, Kelch, B.A. | | Deposit date: | 2022-02-20 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA.
Elife, 11, 2022
|
|
6V1I
 
 | | Cryo-EM reconstruction of the thermophilic bacteriophage P74-26 small terminase- symmetric | | Descriptor: | Small terminase protein | | Authors: | Hayes, J.A, Hilbert, B.J, Gaubitz, C, Stone, N.P, Kelch, B.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A thermophilic phage uses a small terminase protein with a fixed helix-turn-helix geometry.
J.Biol.Chem., 295, 2020
|
|
6VVO
 
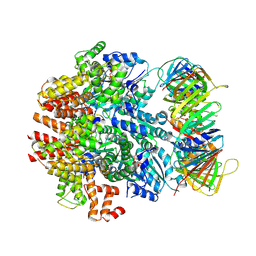 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
4ZNJ
 
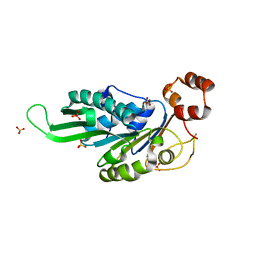 | | Thermus Phage P74-26 Large Terminase ATPase domain mutant R139A (I 2 3 space group) | | Descriptor: | Phage terminase large subunit, SULFATE ION | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZNK
 
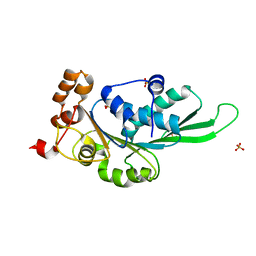 | | Thermus Phage P74-26 Large Terminase ATPase domain from (P 32 2 1 space group) | | Descriptor: | Phage terminase large subunit, SULFATE ION | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZNI
 
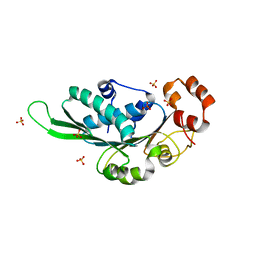 | | Thermus Phage P74-26 Large Terminase ATPase domain (I 2 3 space group) | | Descriptor: | Phage terminase large subunit, SULFATE ION | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Sankaran, B, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZNL
 
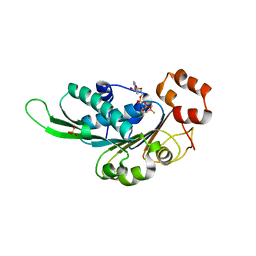 | | Thermus Phage P74-26 Large Terminase ATPase domain bound to ADP Beryllium Fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5E0T
 
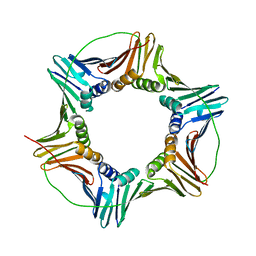 | | Human PCNA mutant - S228I | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Duffy, C.M, Hilbert, B.J, Kelch, B.A. | | Deposit date: | 2015-09-29 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6653 Å) | | Cite: | A Disease-Causing Variant in PCNA Disrupts a Promiscuous Protein Binding Site.
J.Mol.Biol., 428, 2016
|
|
5E0U
 
 | |
5E0V
 
 | |
6BL5
 
 | | Head decoration protein from the hyperthermophilic phage P74-26 | | Descriptor: | Head decoration protein | | Authors: | Stone, N.P, Hilbert, B.J, Hidalgo, D, Halloran, K.T, Kelch, B.A. | | Deposit date: | 2017-11-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Hyperthermophilic Phage Decoration Protein Suggests Common Evolutionary Origin with Herpesvirus Triplex Proteins and an Anti-CRISPR Protein.
Structure, 26, 2018
|
|
7KS4
 
 | |
8UH7
 
 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Primer DNA strand, ... | | Authors: | Gee, C.L, Marcus, K, Kelch, B.A, Makino, D.L. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.628 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
