2DJY
 
 | | Solution structure of Smurf2 WW3 domain-Smad7 PY peptide complex | | Descriptor: | Mothers against decapentaplegic homolog 7, Smad ubiquitination regulatory factor 2 | | Authors: | Chong, P.A, Lin, H, Wrana, J.L, Forman-Kay, J.D. | | Deposit date: | 2006-04-06 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An Expanded WW Domain Recognition Motif Revealed by the Interaction between Smad7 and the E3 Ubiquitin Ligase Smurf2.
J.Biol.Chem., 281, 2006
|
|
2A36
 
 | | Solution structure of the N-terminal SH3 domain of DRK | | Descriptor: | Protein E(sev)2B | | Authors: | Forman-Kay, J.D, Bezsonova, I, Singer, A, Choy, W.-Y, Tollinger, M. | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
2A37
 
 | | Solution structure of the T22G mutant of N-terminal SH3 domain of DRK (DRKN SH3 DOMAIN) | | Descriptor: | Protein E(sev)2B | | Authors: | Bezsonova, I, Singer, A, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
2EZ5
 
 | | Solution Structure of the dNedd4 WW3* Domain- Comm LPSY Peptide Complex | | Descriptor: | Commissureless LPSY Peptide, E3 ubiquitin-protein ligase NEDD4 | | Authors: | Kanelis, V, Bruce, M.C, Skrynnikov, N.R, Rotin, D, Forman-Kay, J.D. | | Deposit date: | 2005-11-10 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants for High-Affinity Binding in a Nedd4 WW3(*) Domain-Comm PY Motif Complex
Structure, 14, 2006
|
|
2K6D
 
 | | CIN85 Sh3-C domain in complex with ubiquitin | | Descriptor: | SH3 domain-containing kinase-binding protein 1, Ubiquitin | | Authors: | Forman-Kay, J, Bezsonova, I. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interactions between the three CIN85 SH3 domains and ubiquitin: implications for CIN85 ubiquitination.
Biochemistry, 47, 2008
|
|
1I5H
 
 | |
2JQZ
 
 | | Solution Structure of the C2 domain of human Smurf2 | | Descriptor: | E3 ubiquitin-protein ligase SMURF2 | | Authors: | Wiesner, S, Ogunjimi, A.A, Wang, H, Rotin, D, Sicheri, F, Wrana, J.L, Forman-Kay, J.D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of the HECT-Type Ubiquitin Ligase Smurf2 through Its C2 Domain
Cell(Cambridge,Mass.), 130, 2007
|
|
2KXQ
 
 | | Solution Structure of Smurf2 WW2 and WW3 bound to Smad7 PY motif containing peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF2, Smad7 PY motif containing peptide | | Authors: | Chong, A, Lin, H, Wrana, J, Forman-Kay, J.D. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LC4
 
 | | Solution Structure of PilP from Pseudomonas aeruginosa | | Descriptor: | PilP protein | | Authors: | Howell, P, Tammam, S, Chong, P, Forman-Kay, J.D. | | Deposit date: | 2011-04-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the PilN, PilO and PilP type IVa pilus subcomplex.
Mol.Microbiol., 82, 2011
|
|
1G0V
 
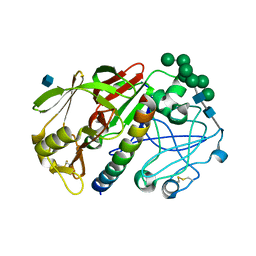 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
1FMX
 
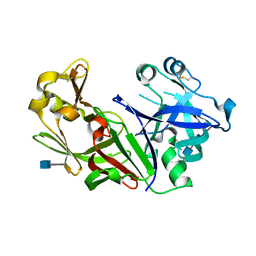 | | STRUCTURE OF NATIVE PROTEINASE A IN THE SPACE GROUP P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1FMU
 
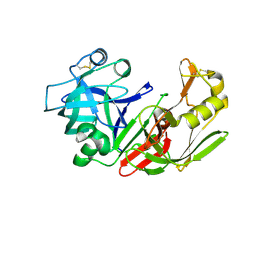 | | STRUCTURE OF NATIVE PROTEINASE A IN P3221 SPACE GROUP. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1DPJ
 
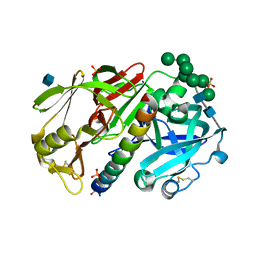 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH IA3 PEPTIDE INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEINASE A, PROTEINASE INHIBITOR IA3 PEPTIDE, ... | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-27 | | Release date: | 2000-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1DP5
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT INHIBITOR | | Descriptor: | PROTEINASE A, PROTEINASE INHIBITOR IA3, beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-23 | | Release date: | 2000-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1K2F
 
 | | siah, Seven In Absentia Homolog | | Descriptor: | BETA-MERCAPTOETHANOL, ZINC ION, siah-1A protein | | Authors: | Polekhina, G, House, C.M, Traficante, N, Mackay, J.P, Relaix, F, Sassoon, D.A, Parker, M.W, Bowtell, D.D.L. | | Deposit date: | 2001-09-26 | | Release date: | 2001-12-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling.
Nat.Struct.Biol., 9, 2002
|
|
3UK3
 
 | | Crystal structure of ZNF217 bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', CHLORIDE ION, ... | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rediscovering DNA recognition by classical Zinc Fingers.
To be Published
|
|
7TO8
 
 | |
7TOA
 
 | |
1XZY
 
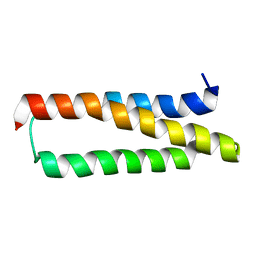 | | Solution structure of the P30-trans form of Alpha Hemoglobin Stabilizing Protein (AHSP) | | Descriptor: | Alpha-hemoglobin stabilizing protein | | Authors: | Gell, D.A, Feng, L, Zhou, S, Kong, Y, Lee, C, Weiss, M.J, Shi, Y, Mackay, J.P. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin
Cell(Cambridge,Mass.), 119, 2004
|
|
1Y03
 
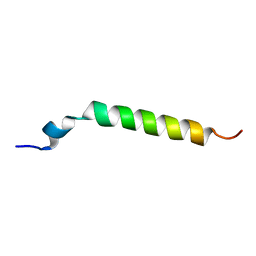 | | Solution structure of a recombinant type I sculpin antifreeze protein | | Descriptor: | Antifreeze peptide SS-3 | | Authors: | Kwan, A.H.Y, Fairley, K, Anderberg, P.I, Liew, C.W, Harding, M.M, Mackay, J.P. | | Deposit date: | 2004-11-14 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a recombinant type I sculpin antifreeze protein
Biochemistry, 44, 2005
|
|
3G9Y
 
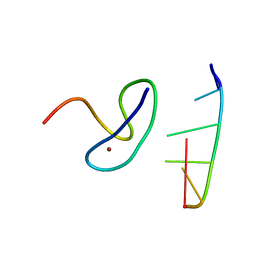 | | Crystal structure of the second zinc finger from ZRANB2/ZNF265 bound to 6 nt ssRNA sequence AGGUAA | | Descriptor: | RNA (5'-R(*AP*GP*GP*UP*AP*A)-3'), ZINC ION, Zinc finger Ran-binding domain-containing protein 2 | | Authors: | Loughlin, F.E, McGrath, A.P, Lee, M, Guss, J.M, Mackay, J.P. | | Deposit date: | 2009-02-15 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7JX7
 
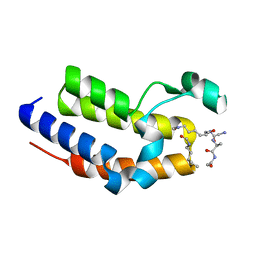 | | BRD2-BD2 in complex with a diacetylated-H2A.Z peptide | | Descriptor: | Bromodomain-containing protein 2, Diacetylated-H2A.Z peptide | | Authors: | Patel, K, Low, J.K.K, Mackay, J.P, Solomon, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The bromodomains of BET family proteins can recognize diacetylated histone H2A.Z.
Protein Sci., 30, 2021
|
|
8TX8
 
 | | Crystal Structure of RBBP4 bound to ZNF512B peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Deshpande, C.N, Mackay, J.P. | | Deposit date: | 2023-08-22 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ZNF512B binds RBBP4 via a variant NuRD interaction motif and aggregates chromatin in a NuRD complex-independent manner.
Nucleic Acids Res., 52, 2024
|
|
8D4Y
 
 | | C-terminal SANT-SLIDE domain of human Chromodomain-helicase-DNA-binding protein 4 (CHD4) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Moghaddas Sani, H, Deshpande, C.N, Panjikar, S, Patel, K, Mackay, J.P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families.
Nat Commun, 13, 2022
|
|
7L4Z
 
 | | Structure of SARS-CoV-2 spike RBD in complex with cyclic peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-DTY-LYS-ALA-GLY-VAL-VAL-TYR-GLY-TYR-ASN-ALA-TRP-ILE-ARG-CYS-NH2, Spike protein S1 | | Authors: | Christie, M, Mackay, J.P, Passioura, T, Payne, R.J. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Discovery of Cyclic Peptide Ligands to the SARS-CoV-2 Spike Protein Using mRNA Display.
Acs Cent.Sci., 7, 2021
|
|
