6J61
 
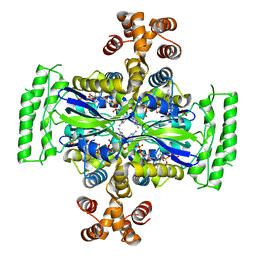 | | Crystal Structure of Thymidylate Synthase, Thy1, from Thermus thermophilus having an Extra C Terminal Domain | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, PHOSPHATE ION | | Authors: | Ogawa, A, Sampei, G, Kawai, G. | | Deposit date: | 2019-01-12 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the flavin-dependent thymidylate synthase Thy1 from Thermus thermophilus with an extra C-terminal domain.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1J1H
 
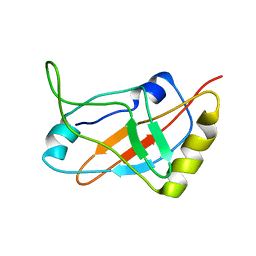 | | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus | | Descriptor: | Small Protein B | | Authors: | Someya, T, Nameki, N, Hosoi, H, Suzuki, S, Hatanaka, H, Fujii, M, Terada, T, Shirouzu, M, Inoue, Y, Shibata, T, Kuramitsu, S, Yokoyama, S, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-04 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus
FEBS Lett., 535, 2003
|
|
1IT1
 
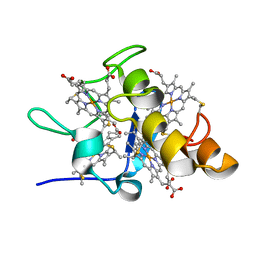 | | Solution structures of ferrocytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | Descriptor: | HEME C, cytochrome c3 | | Authors: | Harada, E, Fukuoka, Y, Ohmura, T, Fukunishi, A, Kawai, G, Fujiwara, T, Akutsu, H. | | Deposit date: | 2001-12-29 | | Release date: | 2002-07-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Redox-coupled conformational alternations in cytochrome c(3) from D. vulgaris Miyazaki F on the basis of its reduced solution structure.
J.Mol.Biol., 319, 2002
|
|
2F87
 
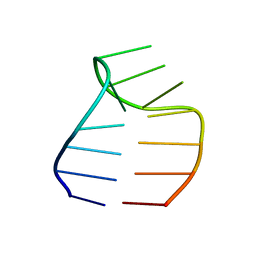 | | Solution structure of a GAAG tetraloop in SRP RNA from Pyrococcus furiosus | | Descriptor: | SRP RNA | | Authors: | Okada, K, Takahashi, M, Sakamoto, T, Nakamura, K, Kanai, A, Kawai, G | | Deposit date: | 2005-12-02 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a GAAG tetraloop in helix 6 of SRP RNA from Pyrococcus furiosus
Nucleosides Nucleotides Nucleic Acids, 25, 2006
|
|
6IZP
 
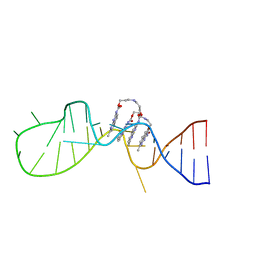 | | Solution structure of the complex of naphthyridine carbamate dimer and an RNA with UGGAA-UGGAA pentad | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, RNA (29-MER) | | Authors: | Nagano, K, Shibata, T, Nakatani, K, Kawai, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun, 12, 2021
|
|
2Z02
 
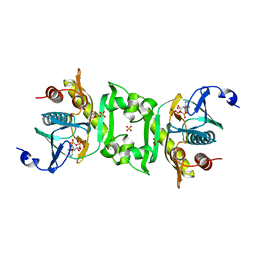 | | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase wit ATP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase from Methanocaldococcus jannaschii
To be Published
|
|
1WKS
 
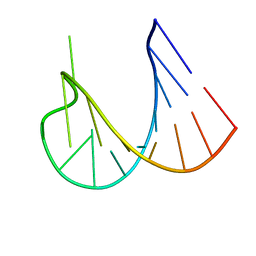 | |
1XWP
 
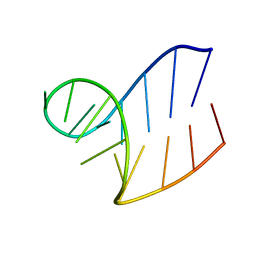 | | Solution structure of AUCGCA loop | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*UP*CP*GP*CP*AP*CP*UP*CP*CP*A)-3' | | Authors: | Sakamoto, T, Oguro, A, Kawai, G, Ohtsu, T, Nakamura, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structures of double loops of an RNA aptamer against mammalian initiation factor 4A
Nucleic Acids Res., 33, 2005
|
|
1XWU
 
 | | Solution structure of ACAUAGA loop | | Descriptor: | 5'-R(*CP*GP*AP*AP*AP*CP*AP*UP*AP*GP*AP*UP*UP*CP*GP*A)-3' | | Authors: | Sakamoto, T, Oguro, A, Kawai, G, Ohtsu, T, Nakamura, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structures of double loops of an RNA aptamer against mammalian initiation factor 4A
Nucleic Acids Res., 33, 2005
|
|
7FGT
 
 | | Discovery of DS15060524; Gene targeting chimera (GeneTAC) for the treatment of Friedreich's Ataxia (FRDA) | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*GP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*TP*CP*TP*T)-3'), ~{N}-[3-[3-(dimethylamino)propylamino]-3-oxidanylidene-propyl]-1-methyl-4-[3-[[1-methyl-4-[[1-methyl-4-[3-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]propanoylamino]imidazol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]propanoylamino]imidazole-2-carboxamide | | Authors: | Takase, N, Kawai, G, Igarashi, W, Katagiri, T. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery of DS15060524; Gene targeting chimera (GeneTAC) for the treatment of Friedreich's Ataxia (FRDA)
To be published
|
|
7EXY
 
 | |
2YYU
 
 | | Crystal structure of uncharacterized conserved protein from Geobacillus kaustophilus | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Kanagawa, M, Baba, S, Nakamura, Y, Bessho, Y, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Geobacillus kaustophilus
To be Published
|
|
2D17
 
 | | Solution RNA structure of stem-bulge-stem region of the HIV-1 dimerization initiation site | | Descriptor: | 5'-R(*CP*GP*GP*CP*AP*AP*GP*AP*GP*GP*CP*GP*AP*CP*CP*C)-3', 5'-R(*GP*GP*GP*UP*CP*GP*GP*CP*UP*UP*GP*CP*UP*G)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D1A
 
 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the extended-duplex dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2D1B
 
 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the kissing-loop dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2RRC
 
 | | Solution Structure of RNA aptamer against AML1 Runt domain | | Descriptor: | 5'-R(P*GP*GP*AP*CP*CP*CP*(AP7)P*CP*CP*AP*CP*GP*GP*CP*GP*AP*GP*GP*UP*CP*CP*A)-3' | | Authors: | Nomura, Y, Fujiwara, K, Chiba, M, Fukunaga, J, Tanaka, Y, Iibuchi, H, Tanaka, T, Nakamura, Y, Kawai, G, Kozu, T, Sakamoto, T. | | Deposit date: | 2010-06-23 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A novel high affinity RNA motif that mimics DNA in AML1 Runt domain binding
To be Published
|
|
7FJ0
 
 | |
7FHI
 
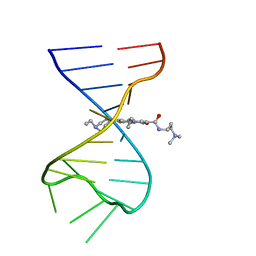 | |
7DD4
 
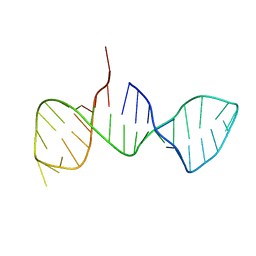 | |
2D19
 
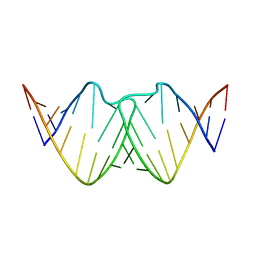 | | Solution RNA structure of loop region of the HIV-1 dimerization initiation site in the kissing-loop dimer | | Descriptor: | 5'-R(*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*GP*GP*C)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2CUW
 
 | | Crystal Structure of Thermus thermophilus PurS, one of the subunits of Formylglycinamide Ribonucleotide Amidotransferase in the purine biosynthetic pathway | | Descriptor: | PurS | | Authors: | Yanai, H, Kanagawa, M, Sampei, G, Kawai, G, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-30 | | Release date: | 2005-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Thermus thermophilus PurS, One of the Subunits of Formylglycinamide Ribonucleotide Amidotransferase in the Purine Biosynthetic Pathway
To be Published
|
|
2D18
 
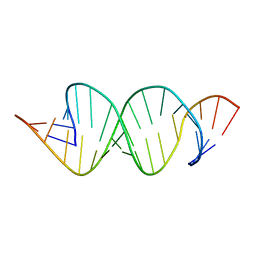 | | Solution RNA structure of loop region of the HIV-1 dimerization initiation site in the extended-duplex dimer | | Descriptor: | 5'-R(*GP*CP*UP*GP*AP*AP*GP*UP*GP*CP*AP*CP*AP*CP*GP*GP*C)-3' | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2FDT
 
 | | Solution structure of a conserved RNA hairpin of eel LINE UnaL2 | | Descriptor: | 36-mer | | Authors: | Nomura, Y, Baba, S, Nakazato, S, Sakamoto, T, Kajikawa, M, Okada, N, Kawai, G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional importance of a conserved RNA hairpin of eel LINE UnaL2
Nucleic Acids Res., 34, 2006
|
|
2YW2
 
 | | Crystal structure of GAR synthetase from Aquifex aeolicus in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, Phosphoribosylamine--glycine ligase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-19 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
2YRX
 
 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
