4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | Descriptor: | Major vault protein | | Authors: | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2008-10-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
4MKP
 
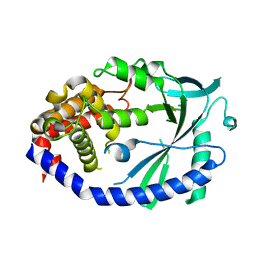 | | Crystal structure of human cGAS apo form | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS
Plos One, 8, 2013
|
|
5JYJ
 
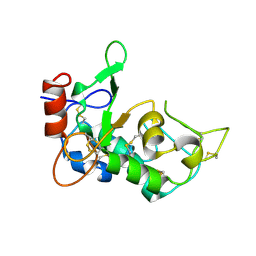 | | Crystal structure of mouse JUNO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sperm-egg fusion protein Juno | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-05-14 | | Release date: | 2017-05-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the egg IZUMO1 receptor JUNO
To Be Published
|
|
7Y9Y
 
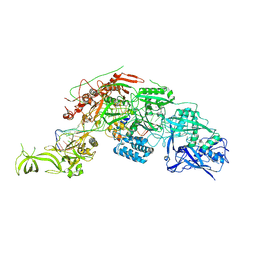 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (no PFS) complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, RNA (27-MER), ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7Y9X
 
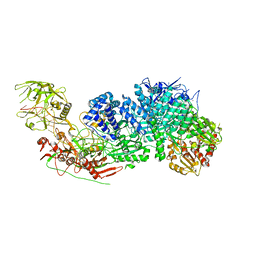 | | Structure of the Cas7-11-Csx29-guide RNA complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, ZINC ION, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-06-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
8WEY
 
 | | PSI-LHCI of the red alga Cyanidium caldarium RK-1 (NIES-2137) | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{R} )-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Kato, K, Hamaguchi, T, Nakajima, Y, Kawakami, K, Yonekura, K, Shen, J.R, Nagao, R. | | Deposit date: | 2023-09-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | The structure of PSI-LHCI from Cyanidium caldarium provides evolutionary insights into conservation and diversity of red-lineage LHCs.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8GS2
 
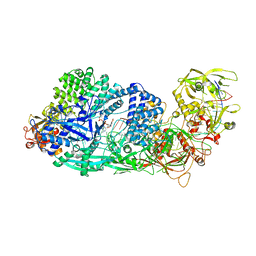 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching PFS) complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHAT domain-containing protein, CRISPR-associated RAMP family protein, ... | | Authors: | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
7XHT
 
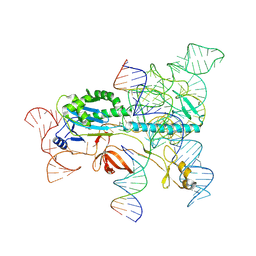 | | Structure of the OgeuIscB-omega RNA-target DNA complex | | Descriptor: | DNA (49-MER), DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3'), LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Kato, K, Okazaki, O, Isayama, Y, Ishikawa, J, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2022-04-10 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structure of the IscB-omega RNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9.
Nat Commun, 13, 2022
|
|
8XLS
 
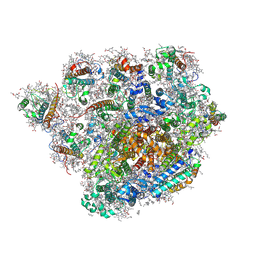 | | PSI-FCPI of the diatom Thalassiosira pseudonana CCMP1335 | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{R} )-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Kato, K, Nakajima, Y, Shen, J.R, Nagao, R. | | Deposit date: | 2023-12-26 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for molecular assembly of fucoxanthin chlorophyll a/c-binding proteins in a diatom photosystem I supercomplex
Elife, 2024
|
|
2KP2
 
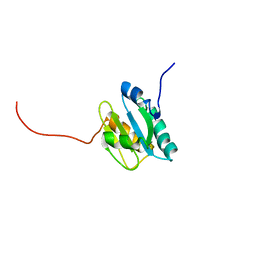 | |
2KP1
 
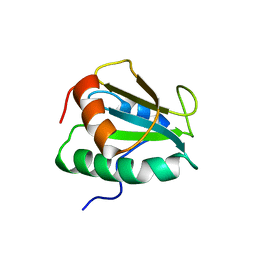 | |
7JL4
 
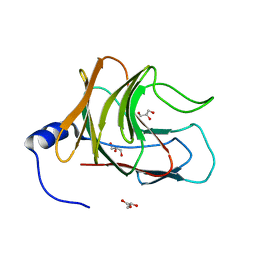 | | Crystal structure of TRIM65 PSpry domain | | Descriptor: | GLYCEROL, Tripartite motif-containing protein 65 | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL3
 
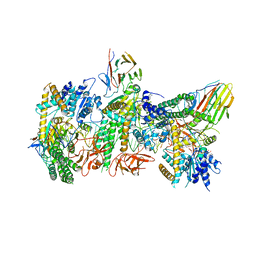 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL0
 
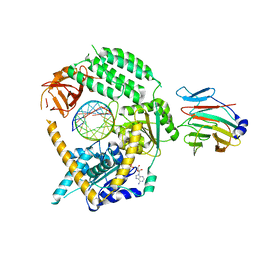 | | Cryo-EM structure of MDA5-dsRNA in complex with TRIM65 PSpry domain (Monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL1
 
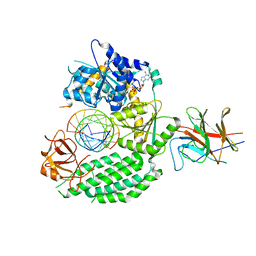 | | Cryo-EM structure of RIG-I:dsRNA in complex with RIPLET PrySpry domain (monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL2
 
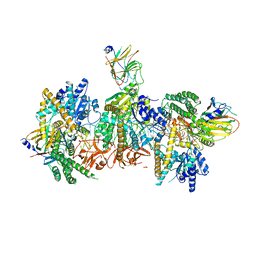 | | Cryo-EM structure of MDA5-dsRNA filament in complex with TRIM65 PSpry domain (Trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
4GTW
 
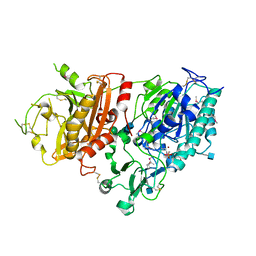 | | Crystal structure of mouse Enpp1 in complex with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GTZ
 
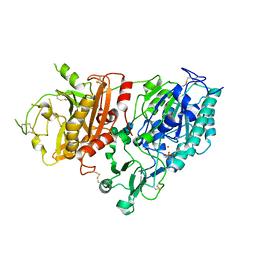 | | Crystal structure of mouse Enpp1 in complex with CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GTY
 
 | | Crystal structure of mouse Enpp1 in complex with GMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GTX
 
 | | Crystal structure of mouse Enpp1 in complex with TMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4XJ6
 
 | | Crystal structure of Escherichia coli DncV 3'-deoxy GTP bound form | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, VC0179-like protein | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ3
 
 | | Crystal structure of Vibrio cholerae DncV GTP bound form | | Descriptor: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ1
 
 | | Crystal structure of Vibrio cholerae DncV apo form | | Descriptor: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ4
 
 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy ATP bound form | | Descriptor: | 1,2-ETHANEDIOL, 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
4XJ5
 
 | | Crystal structure of Vibrio cholerae DncV 3'-deoxy GTP bound form | | Descriptor: | 1,2-ETHANEDIOL, 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, Cyclic AMP-GMP synthase, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
