1EVL
 
 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH A THREONYL ADENYLATE ANALOG | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
2TEP
 
 | | PEANUT LECTIN COMPLEXED WITH T-ANTIGENIC DISACCHARIDE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Ravindran, M, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-04-05 | | Release date: | 1999-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Specificity of Peanut Agglutinin for Thomsen-Friedenreich Antigen is Mediated by Water-Bridges
Curr.Sci., 72, 1997
|
|
3D2C
 
 | |
3D2B
 
 | |
8EXY
 
 | |
8EOF
 
 | |
8EHQ
 
 | |
6QMR
 
 | | Complement factor D in complex with the inhibitor (S)-2-(2-((3'-(1-amino-2-hydroxyethyl)-[1,1'-biphenyl]-3-yl)methoxy)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-[(1~{S})-1-azanyl-2-oxidanyl-ethyl]phenyl]phenyl]methoxy]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
6QMT
 
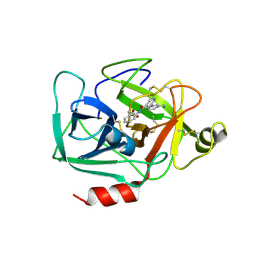 | | Complement factor D in complex with the inhibitor 2-(2-(3'-(aminomethyl)-[1,1'-biphenyl]-3-carboxamido)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
8EOS
 
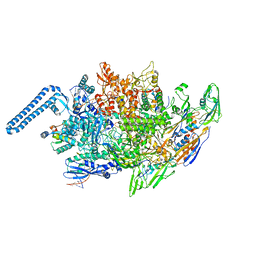 | | M. tuberculosis RNAP elongation complex with NusG and CMPCPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (36-MER), DNA (38-MER), ... | | Authors: | Vishwakarma, R.K, Murakami, K.S. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Allosteric mechanism of transcription inhibition by NusG-dependent pausing of RNA polymerase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EJ3
 
 | |
8EOT
 
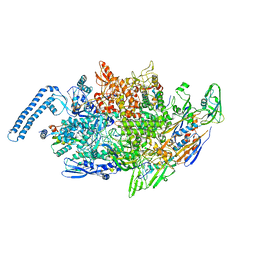 | | M. tuberculosis RNAP elongation complex with NusG | | Descriptor: | DNA (31-MER), DNA (33-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Vishwakarma, R.K, Murakami, K.S. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric mechanism of transcription inhibition by NusG-dependent pausing of RNA polymerase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UXE
 
 | | Pseudomonas phage E217 small terminase (TerS) | | Descriptor: | Small terminase | | Authors: | Lokareddy, R.K, Hou, C.-F.D, Doll, S.G, Li, F, Gillilan, R, Forti, F, Briani, F, Cingolani, G. | | Deposit date: | 2022-05-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Terminase Subunits from the Pseudomonas-Phage E217.
J.Mol.Biol., 434, 2022
|
|
8EOE
 
 | |
7UEB
 
 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO2) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
7UEA
 
 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO1) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
1JAC
 
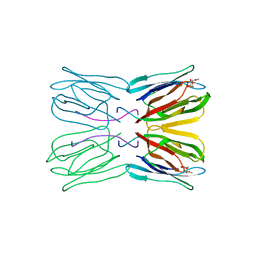 | | A NOVEL MODE OF CARBOHYDRATE RECOGNITION IN JACALIN, A MORACEAE PLANT LECTIN WITH A BETA-PRISM | | Descriptor: | JACALIN, methyl alpha-D-galactopyranoside | | Authors: | Sankaranarayanan, R, Sekar, K, Banerjee, R, Sharma, V, Surolia, A, Vijayan, M. | | Deposit date: | 1996-05-22 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A novel mode of carbohydrate recognition in jacalin, a Moraceae plant lectin with a beta-prism fold.
Nat.Struct.Biol., 3, 1996
|
|
4PVZ
 
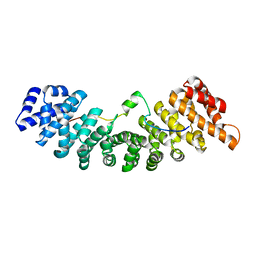 | | Structure of yeast importin a bound to the membrane protein Nuclear Localization Signal sequence of INM protein Heh2 | | Descriptor: | Importin subunit alpha, Inner nuclear membrane protein HEH2 | | Authors: | Lokareddy, R.K, Hapsari, A.R, van Rheenen, M, Bhardwaj, A, Veenhoff, L.M, Cingolani, C. | | Deposit date: | 2014-03-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distinctive Properties of the Nuclear Localization Signals of Inner Nuclear Membrane Proteins Heh1 and Heh2.
Structure, 23, 2015
|
|
3IT4
 
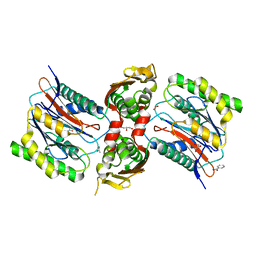 | | The Crystal Structure of Ornithine Acetyltransferase from Mycobacterium tuberculosis (Rv1653) at 1.7 A | | Descriptor: | ACETATE ION, Arginine biosynthesis bifunctional protein argJ alpha chain, Arginine biosynthesis bifunctional protein argJ beta chain, ... | | Authors: | Sankaranarayanan, R, Cherney, M.M, Garen, C, Garen, G, Yuan, M, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-08-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of ornithine acetyltransferase from Mycobacterium tuberculosis bound to ornithine, a competitive inhibitor.
J.Mol.Biol., 397, 2010
|
|
4HRF
 
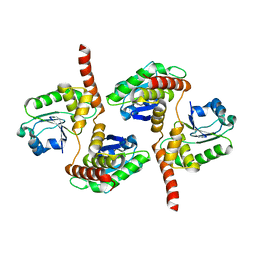 | | Atomic structure of DUSP26 | | Descriptor: | Dual specificity protein phosphatase 26 | | Authors: | Lokareddy, R.K, Bhardwaj, A, Cingolani, G. | | Deposit date: | 2012-10-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Atomic structure of dual-specificity phosphatase 26, a novel p53 phosphatase.
Biochemistry, 52, 2013
|
|
4XZR
 
 | |
1FYF
 
 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE COMPLEXED WITH A SERYL ADENYLATE ANALOG | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2000-09-29 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem.
Cell(Cambridge,Mass.), 103, 2000
|
|
1QF3
 
 | | PEANUT LECTIN COMPLEXED WITH METHYL-BETA-GALACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-04-06 | | Release date: | 1999-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the complexes of peanut lectin with methyl-beta-galactose and N-acetyllactosamine and a comparative study of carbohydrate binding in Gal/GalNAc-specific legume lectins.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5JJ3
 
 | |
2I6U
 
 | | Crystal Structure of Ornithine Carbamoyltransferase complexed with Carbamoyl Phosphate and L-Norvaline from Mycobacterium tuberculosis (Rv1656) at 2.2 A | | Descriptor: | NORVALINE, Ornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Sankaranarayanan, R, Moradian, F, Cherney, L.T, Garen, C, Cherney, M.M, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of ornithine carbamoyltransferase from Mycobacterium tuberculosis and its ternary complex with carbamoyl phosphate and L-norvaline reveal the enzyme's catalytic mechanism
J.Mol.Biol., 375, 2008
|
|
