3MCD
 
 | | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor | | Descriptor: | Cell division topological specificity factor | | Authors: | Kang, G.B, Song, H.E, Kim, M.K, Youn, H.S, Lee, J.G, An, J.Y, Jeon, H, Chun, J.S, Eom, S.H. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor
Mol.Microbiol., 76, 2010
|
|
3G8E
 
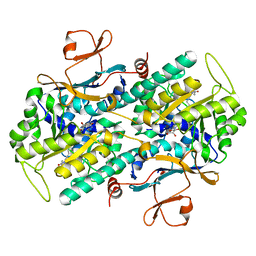 | | Crystal Structure of Rattus norvegicus Visfatin/PBEF/Nampt in Complex with an FK866-based inhibitor | | Descriptor: | 3-[(1E)-3-oxo-3-({4-[1-(phenylcarbonyl)piperidin-4-yl]butyl}amino)prop-1-en-1-yl]-1-beta-D-ribofuranosylpyridinium, Nicotinamide phosphoribosyltransferase | | Authors: | Kang, G.B, Bae, M.H, Kim, M.K, Im, I, Kim, Y.C, Eom, S.H. | | Deposit date: | 2009-02-12 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Rattus norvegicus Visfatin/PBEF/Nampt in complex with an FK866-based inhibitor
Mol.Cells, 27, 2009
|
|
1XNH
 
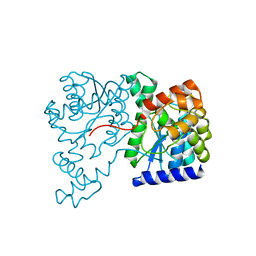 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
1XNG
 
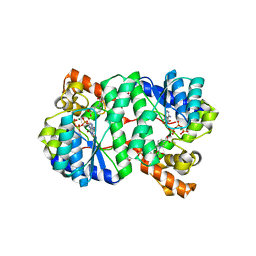 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
5AYX
 
 | | Crystal structure of Human Quinolinate Phosphoribosyltransferase | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Kang, G.B, Kim, M.-K, Im, Y.J, Lee, J.H, Youn, H.-S, An, J.Y, Lee, J.-G, Fukuoka, S.-I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
3K9O
 
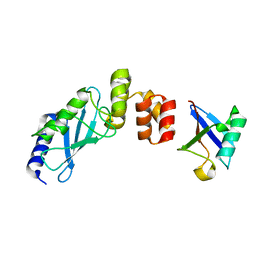 | | The crystal structure of E2-25K and UBB+1 complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of E2-25K/UBB+1 Interaction for Neurotoxicity of Alzheimer Disease by Proteasome Inhibition
To be Published
|
|
3K9P
 
 | | The crystal structure of E2-25K and ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | Authors: | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | Deposit date: | 2009-10-16 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of E2-25K/UBB+1 interaction leading to proteasome inhibition and neurotoxicity
J.Biol.Chem., 285, 2010
|
|
3KU7
 
 | | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor | | Descriptor: | Cell division topological specificity factor | | Authors: | Kang, G.B, Song, H.E, Kim, M.K, Eom, S.H. | | Deposit date: | 2009-11-26 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Helicobacter pylori MinE, a cell division topological specificity factor
Mol.Microbiol., 76, 2010
|
|
3L4F
 
 | | Crystal Structure of betaPIX Coiled-Coil Domain and Shank PDZ Complex | | Descriptor: | Rho guanine nucleotide exchange factor 7, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Im, Y.J, Kang, G.B, Lee, J.H, Song, H.E, Park, K.R, Kim, E, Song, W.K, Park, D, Eom, S.H. | | Deposit date: | 2009-12-19 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for asymmetric association of the betaPIX coiled coil and shank PDZ
J.Mol.Biol., 397, 2010
|
|
2Z3B
 
 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
2Z3A
 
 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
4L1C
 
 | | Crystal structure of Dimerized N-terminal Domain of MinC | | Descriptor: | Probable septum site-determining protein MinC | | Authors: | An, J.Y, Kim, T.G, Park, K.R, Lee, J.G, Youn, H.S, Kang, J.Y, Lee, Y, Kang, G.B, Eom, S.H. | | Deposit date: | 2013-06-03 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the N-terminal domain of MinC dimerized via domain swapping.
J Synchrotron Radiat, 20, 2013
|
|
4Q0F
 
 | | Crystal Structure of Thermotoga maritima FtsH Periplasmic domain | | Descriptor: | ATP-dependent zinc metalloprotease FtsH | | Authors: | An, J.Y, Sharif, H, Barrera, F.N, Karabadzhak, A, Kang, G.B, Park, K.J, Sakkiah, S, Lee, K.W, Lee, S, Engelman, D.M, Wang, J, Eom, S.H. | | Deposit date: | 2014-04-01 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structures of periplasmic and transmembrane domains of FtsH suggest a reverse translocon mechanism for protein extraction from membrane
To be Published
|
|
1XHK
 
 | | Crystal structure of M. jannaschii Lon proteolytic domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative protease La homolog, SULFATE ION | | Authors: | Im, Y.J, Na, Y, Kang, G.B, Rho, S.-H, Kim, M.-K, Lee, J.H, Chung, C.H, Eom, S.H. | | Deposit date: | 2004-09-20 | | Release date: | 2004-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The active site of a lon protease from Methanococcus jannaschii distinctly differs from the canonical catalytic Dyad of Lon proteases.
J.Biol.Chem., 279, 2004
|
|
5AYZ
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE PRODUCT NICOTINATE MONONUCLEOTIDE | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
2R65
 
 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division protease ftsH homolog | | Authors: | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
2R62
 
 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH | | Descriptor: | Cell division protease ftsH homolog | | Authors: | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | Deposit date: | 2007-09-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
2B8I
 
 | | Crystal Structure and Functional Studies Reveal that PAS Factor from Vibrio vulnificus is a Novel Member of the Saposin-Fold Family | | Descriptor: | PAS factor | | Authors: | Lee, J.H, Yang, S.T, Rho, S.H, Im, Y.J, Kim, S.Y, Kim, Y.R, Kim, M.K, Kang, G.B, Kim, J.I, Rhee, J.H, Eom, S.H. | | Deposit date: | 2005-10-07 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and functional studies reveal that PAS factor from Vibrio vulnificus is a novel member of the saposin-fold family
J.Mol.Biol., 355, 2006
|
|
2FJK
 
 | | Crystal structure of Fructose-1,6-Bisphosphate Aldolase in Thermus caldophilus | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase | | Authors: | Lee, J.H, Im, Y.J, Rho, S.-H, Kim, M.-K, Kang, G.B, Eom, S.H. | | Deposit date: | 2006-01-03 | | Release date: | 2006-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stereoselectivity of fructose-1,6-bisphosphate aldolase in Thermus caldophilus
Biochem.Biophys.Res.Commun., 347, 2006
|
|
1N7F
 
 | | Crystal structure of the sixth PDZ domain of GRIP1 in complex with liprin C-terminal peptide | | Descriptor: | 8-mer peptide from interacting protein (liprin), AMPA receptor interacting protein GRIP | | Authors: | Im, Y.J, Park, S.H, Rho, S.H, Lee, J.H, Kang, G.B, Sheng, M, Kim, E, Eom, S.H. | | Deposit date: | 2002-11-14 | | Release date: | 2003-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization
J.BIOL.CHEM., 278, 2003
|
|
4M8A
 
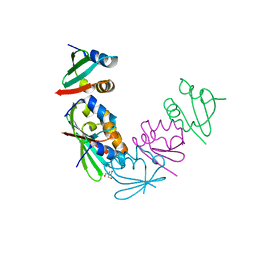 | | Crystal Structure of Thermotoga maritima FtsH Periplasmic Domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-dependent zinc metalloprotease FtsH | | Authors: | An, J.Y, Sharif, H, Barrera, F.N, Karabadzhak, A, Kang, G.B, Park, K.J, Sakkiah, S, Lee, K.W, Lee, S, Engelman, D.M, Wang, J, Eom, S.H. | | Deposit date: | 2013-08-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural roles of periplasmic and transmembrane domains of FtsH in ATP-dependent proteolysis
To be Published
|
|
1N7E
 
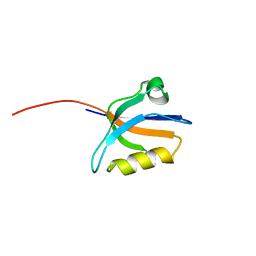 | | Crystal structure of the sixth PDZ domain of GRIP1 | | Descriptor: | AMPA receptor interacting protein GRIP | | Authors: | Im, Y.J, Park, S.H, Rho, S.H, Lee, J.H, Kang, G.B, Sheng, M, Kim, E, Eom, S.H. | | Deposit date: | 2002-11-14 | | Release date: | 2003-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization
J.BIOL.CHEM., 278, 2003
|
|
1Q3O
 
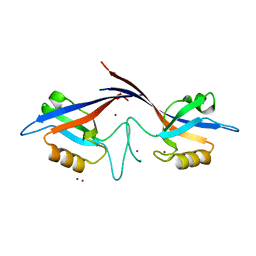 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | Descriptor: | BROMIDE ION, Shank1 | | Authors: | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
1Q3P
 
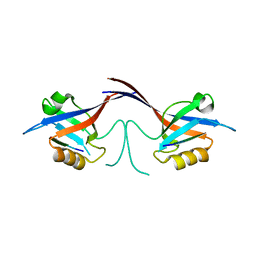 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | Descriptor: | C-terminal hexapeptide from Guanylate kinase-associated protein, Shank1 | | Authors: | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
