6UF8
 
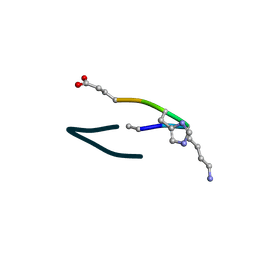 | | S2 symmetric peptide design number 6, London Bridge | | Descriptor: | S2-6, London Bridge | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UFU
 
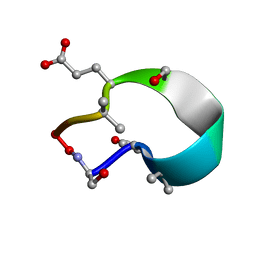 | | C2 symmetric peptide design number 1, Zappy, crystal form 1 | | Descriptor: | C2-1, Zappy, crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UG3
 
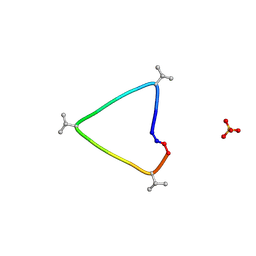 | | C3 symmetric peptide design number 1, Sporty, crystal form 1 | | Descriptor: | C3-1, Sporty, crystal form 1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UGC
 
 | | C3 symmetric peptide design number 3 | | Descriptor: | C3-3 cyclic peptide design, CADMIUM ION, SODIUM ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDR
 
 | | S2 symmetric peptide design number 3 crystal form 1, Lurch | | Descriptor: | S2-3, Lurch crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
4G0L
 
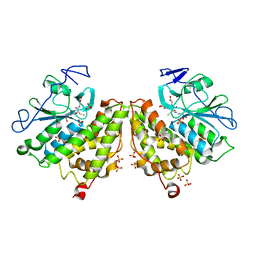 | | Glutathionyl-hydroquinone Reductase, YqjG, of E.coli complexed with GSH | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
6VBY
 
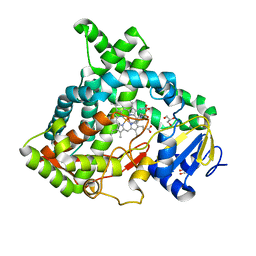 | | Cinnamate 4-hydroxylase (C4H1) from Sorghum bicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cinnamic acid 4-hydroxylase, GLYCEROL, ... | | Authors: | Zhang, B, Kang, C, Lewis, K.M. | | Deposit date: | 2019-12-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of the Cytochrome P450 Monooxygenase Cinnamate 4-hydroxylase fromSorghum bicolor.
Plant Physiol., 183, 2020
|
|
1SMM
 
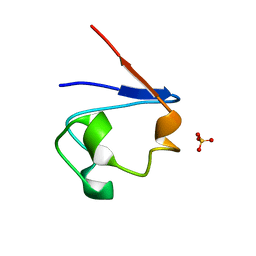 | | Crystal Structure of Cp Rd L41A mutant in oxidized state | | Descriptor: | FE (III) ION, Rubredoxin, SULFATE ION | | Authors: | Park, I.Y, Youn, B, Harley, J.L, Eidsness, M.K, Smith, E, Ichiye, T, Kang, C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer
J.BIOL.INORG.CHEM., 9, 2004
|
|
1SM5
 
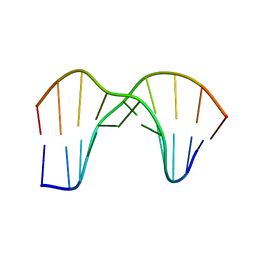 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*(BRU)P*TP*AP*AP*TP*(BRU)P*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2004-03-08 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SMU
 
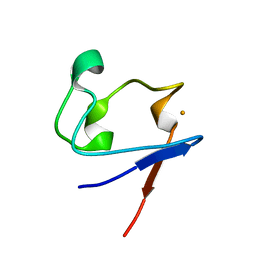 | | Crystal Structure of Cp Rd L41A mutant in reduced state 1 (drop-reduced) | | Descriptor: | FE (II) ION, Rubredoxin | | Authors: | Park, I.Y, Youn, B, Harley, J.L, Eidsness, M.K, Smith, E, Ichiye, T, Kang, C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer
J.BIOL.INORG.CHEM., 9, 2004
|
|
1T9Q
 
 | | Crystal Structure of V44L Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
1T9P
 
 | | Crystal Structure of V44A, G45P Cp Rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Park, I.Y, Eidsness, M.K, Lin, I.J, Gebel, E.B, Youn, B, Harley, J.L, Machonkin, T.E, Frederick, R.O, Markley, J.L, Smith, E.T, Ichiye, T, Kang, C. | | Deposit date: | 2004-05-18 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic studies of V44 mutants of Clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential.
Proteins, 57, 2004
|
|
1N4E
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*AP*TP*TP*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a DNA decamer containing a cis-syn thymine dimer.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2IUK
 
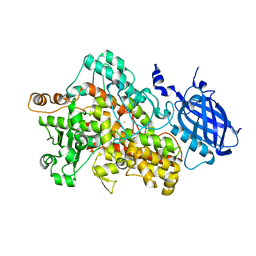 | | Crystal structure of Soybean Lipoxygenase-D | | Descriptor: | FE (III) ION, SEED LIPOXYGENASE | | Authors: | Youn, B, Sellhorn, G.E, Mirchel, R.J, Gaffney, B.J, Grimes, H.D, Kang, C. | | Deposit date: | 2006-06-05 | | Release date: | 2006-10-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Vegetative Soybean Lipoxygenase Vlx-B and Vlx-D, and Comparisons with Seed Isoforms Lox-1 and Lox-3.
Proteins, 65, 2006
|
|
2IUJ
 
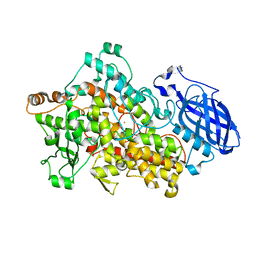 | | Crystal Structure of Soybean Lipoxygenase-B | | Descriptor: | FE (III) ION, LIPOXYGENASE L-5 | | Authors: | Youn, B, Sellhorn, G.E, Mirchel, R.J, Gaffney, B.J, Grimes, H.D, Kang, C. | | Deposit date: | 2006-06-05 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Vegetative Soybean Lipoxygenase Vlx-B and Vlx-D, and Comparisons with Seed Isoforms Lox-1 and Lox-3.
Proteins, 65, 2006
|
|
2LAT
 
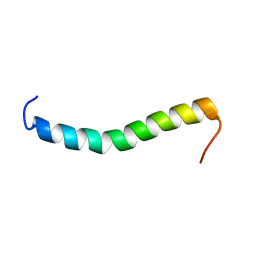 | | Solution structure of a Human minimembrane protein OST4 | | Descriptor: | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 4 | | Authors: | Gayen, S, Kang, C. | | Deposit date: | 2011-03-21 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a human minimembrane protein Ost4, a subunit of the oligosaccharyltransferase complex.
Biochem.Biophys.Res.Commun., 409, 2011
|
|
2M0S
 
 | |
2MFR
 
 | |
2MV6
 
 | |
2ONG
 
 | | Crystal Structure of of limonene synthase with 2-fluorogeranyl diphosphate (FGPP). | | Descriptor: | (1S)-1-[(1R)-1-FLUOROETHYL]-1,5-DIMETHYLHEXYL TRIHYDROGEN DIPHOSPHATE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4S-limonene synthase, ... | | Authors: | Hyatt, D.C, Youn, B, Croteau, R, Kang, C. | | Deposit date: | 2007-01-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of limonene synthase, a simple model for terpenoid cyclase catalysis.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ONH
 
 | | Crystal Structure of of limonene synthase with 2-fluorolinalyl diphosphate(FLPP) | | Descriptor: | (1S)-1-[(1S)-1-FLUOROETHYL]-1,5-DIMETHYLHEXYL TRIHYDROGEN DIPHOSPHATE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4S-limonene synthase, ... | | Authors: | Hyatt, D.C, Youn, B, Croteau, R, Kang, C. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of limonene synthase, a simple model for terpenoid cyclase catalysis.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2NDJ
 
 | | Structural Basis for KCNE3 and Estrogen Modulation of the KCNQ1 Channel | | Descriptor: | Potassium voltage-gated channel subfamily E member 3 | | Authors: | Sanders, C.R, Van Horn, W.D, Kroncke, B.M, Sisco, N.J, Meiler, J, Vanoye, C.G, Song, Y, Nannemann, D.P, Welch, R.C, Kang, C, Smith, J, George, A.L. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for KCNE3 modulation of potassium recycling in epithelia.
Sci Adv, 2, 2016
|
|
2N7G
 
 | |
2N7Q
 
 | |
2MXB
 
 | |
