2D8N
 
 | | Crystal structure of human recoverin at 2.2 A resolution | | Descriptor: | Recoverin | | Authors: | Kamo, S, Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-07 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human recoverin at 2.2 A resolution
To be Published
|
|
2DC5
 
 | | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution | | Descriptor: | Glutathione S-transferase, mu 7 | | Authors: | Kamo, S, Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-28 | | Release date: | 2006-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of mouse glutathione S-transferase, mu7 (GSTM7) at 1.6 A resolution
To be Published
|
|
5N4Q
 
 | | Human myelin protein P2, mutant T51P | | Descriptor: | D-MALATE, Myelin P2 protein, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Kursula, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.
Sci Rep, 7, 2017
|
|
5N4M
 
 | | Human myelin protein P2, mutant I43N | | Descriptor: | D-MALATE, Myelin P2 protein, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Kursula, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.
Sci Rep, 7, 2017
|
|
5N4P
 
 | | Human myelin P2, mutant I52T | | Descriptor: | D-MALATE, Myelin P2 protein, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Kursula, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Molecular mechanisms of Charcot-Marie-Tooth neuropathy linked to mutations in human myelin protein P2.
Sci Rep, 7, 2017
|
|
6STS
 
 | | Human myelin protein P2 mutant R30Q | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2019-09-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XUA
 
 | | Human myelin protein P2 mutant K21Q | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XW9
 
 | | Human myelin protein P2 mutant K120S | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XVQ
 
 | | Human myelin protein P2 mutant K31Q | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XVY
 
 | | Human myelin protein P2 mutant R88Q | | Descriptor: | CHLORIDE ION, CITRIC ACID, Myelin P2 protein, ... | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XU9
 
 | | Human myelin protein P2 mutant K3N | | Descriptor: | CHLORIDE ION, CITRIC ACID, Myelin P2 protein, ... | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XU5
 
 | | Human myelin protein P2 mutant N2D | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XUW
 
 | | Human myelin protein P2 mutant L27D | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XVR
 
 | | Human myelin protein P2 mutant L35S | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
2WFN
 
 | | Filamin A actin binding domain | | Descriptor: | FILAMIN-A, SULFATE ION | | Authors: | Ruskamo, S, Ylanne, J. | | Deposit date: | 2009-04-09 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Human Filamin a Actin-Binding Domain
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6XVS
 
 | | Human myelin protein P2 mutant P38G, unliganded | | Descriptor: | GLYCEROL, Myelin P2 protein | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
2W0P
 
 | |
4BVM
 
 | | The peripheral membrane protein P2 from human myelin at atomic resolution | | Descriptor: | CITRIC ACID, MYELIN P2 PROTEIN, PALMITIC ACID, ... | | Authors: | Ruskamo, S, Yadav, R.P, Kursula, P. | | Deposit date: | 2013-06-26 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Atomic Resolution View Into the Structure-Function Relationships of the Human Myelin Peripheral Membrane Protein P2
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8WY1
 
 | | The structure of cyclization domain in cyclic beta-1,2-glucan synthase from Thermoanaerobacter italicus | | Descriptor: | Glycosyltransferase 36 | | Authors: | Tanaka, N, Saito, R, Kobayashi, K, Nakai, H, Kamo, S, Kuramochi, K, Taguchi, H, Nakajima, M, Masaike, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Functional and structural analysis of a cyclization domain in a cyclic beta-1,2-glucan synthase.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
3ZR8
 
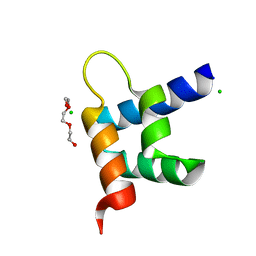 | | Crystal structure of RxLR effector Avr3a11 from Phytophthora capsici | | Descriptor: | AVR3A11, CHLORIDE ION, TRIETHYLENE GLYCOL | | Authors: | Boutemy, L.S, King, S.R.F, Win, J, Hughes, R.K, Clarke, T.A, Blumenschein, T.M.A, Kamoun, S, Banfield, M.J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Structures of Phytophthora Rxlr Effector Proteins: A Conserved But Adaptable Fold Underpins Functional Diversity.
J.Biol.Chem., 286, 2011
|
|
9FP6
 
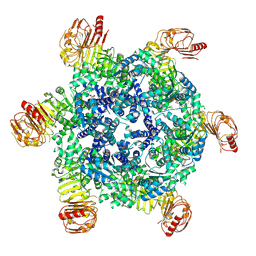 | |
7BNT
 
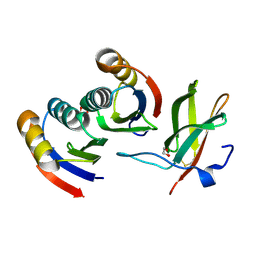 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with a predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Descriptor: | 1,2-ETHANEDIOL, AVR-Pik protein, Predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Authors: | Bialas, A, Langner, T, Harant, A, Contreras, M.P, Stevenson, C.E.M, Lawson, D.M, Sklenar, J, Kellner, R, Moscou, M.J, Terauchi, R, Banfield, M.J, Kamoun, S. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Two NLR immune receptors acquired high-affinity binding to a fungal effector through convergent evolution of their integrated domain.
Elife, 10, 2021
|
|
6ZMM
 
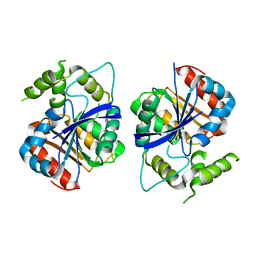 | | Crystal structure of human NDRG1 | | Descriptor: | CHLORIDE ION, Protein NDRG1 | | Authors: | Mustonen, V, Kursula, P, Ruskamo, S. | | Deposit date: | 2020-07-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal and solution structure of NDRG1, a membrane-binding protein linked to myelination and tumour suppression.
Febs J., 288, 2021
|
|
6EXJ
 
 | | PDZ domain from rat Shank3 bound to the C terminus of somatostatin receptor subtype 2 | | Descriptor: | SH3 and multiple ankyrin repeat domains protein 3, SSTR2 | | Authors: | Ponna, S.K, Keller, C, Ruskamo, S, Myllykoski, M, Boeckers, T.M, Kursula, P. | | Deposit date: | 2017-11-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for PDZ domain interactions in the post-synaptic density scaffolding protein Shank3.
J. Neurochem., 145, 2018
|
|
8RFH
 
 | |
