1QR5
 
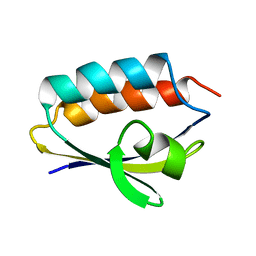 | | SOLUTION STRUCTURE OF HISTIDINE CONTAINING PROTEIN (HPR) FROM STAPHYLOCOCCUS CARNOSUS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Kalbitzer, H.R, Gorler, A, Li, H, Dubovskii, P.V, Hengstenberg, W, Kowolik, C, Yamada, H, Akasaka, K. | | Deposit date: | 1999-05-19 | | Release date: | 2000-06-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | 15N and 1H NMR study of histidine containing protein (HPr) from Staphylococcus carnosus at high pressure.
Protein Sci., 9, 2000
|
|
2KLE
 
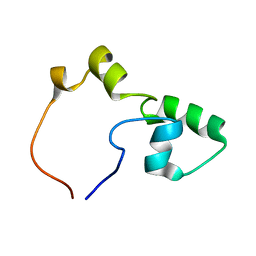 | |
2KLD
 
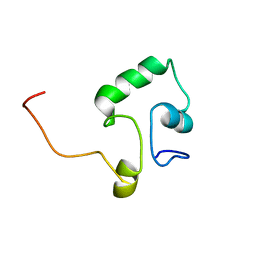 | |
1TXE
 
 | | Solution structure of the active-centre mutant Ile14Ala of the histidine-containing phosphocarrier protein (HPr) from Staphylococcus carnosus | | Descriptor: | Phosphocarrier protein HPr | | Authors: | Moeglich, A, Koch, B, Hengstenberg, W, Brunner, E, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-07-04 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the active-centre mutant I14A of the histidine-containing phosphocarrier protein from Staphylococcus carnosus
Eur.J.Biochem., 271, 2004
|
|
1KA5
 
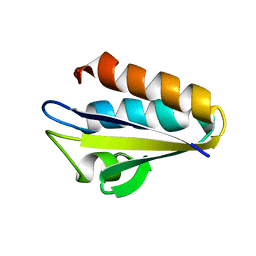 | | Refined Solution Structure of Histidine Containing Phosphocarrier Protein from Staphyloccocus aureus | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Maurer, T, Meier, S, Hengstenberg, W, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2001-10-31 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of the histidine-containing phosphocarrier protein (HPr) from Staphylococcus aureus and characterization of its interaction with the bifunctional HPr kinase/phosphorylase
J.Bacteriol., 186, 2004
|
|
4BGC
 
 | |
1I35
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF THE PROTEIN KINASE BYR2 FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | PROTEIN KINASE BYR2 | | Authors: | Gronwald, W, Huber, F, Grunewald, P, Sporner, M, Wohlgemuth, S, Herrmann, C, Kalbitzer, H.R. | | Deposit date: | 2001-02-13 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras binding domain of the protein kinase Byr2 from Schizosaccharomyces pombe.
Structure, 9, 2001
|
|
1XKE
 
 | | Solution structure of the second Ran-binding domain from human RanBP2 | | Descriptor: | Ran-binding protein 2 | | Authors: | Geyer, J.P, Doeker, R, Kremer, W, Zhao, X, Kuhlmann, J, Kalbitzer, H.R. | | Deposit date: | 2004-09-28 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ran-binding domain 2 of RanBP2 and its interaction with the C terminus of Ran.
J.Mol.Biol., 348, 2005
|
|
1ZTO
 
 | | INACTIVATION GATE OF POTASSIUM CHANNEL RCK4, NMR, 8 STRUCTURES | | Descriptor: | POTASSIUM CHANNEL PROTEIN RCK4 | | Authors: | Antz, C, Geyer, M, Fakler, B, Schott, M, Frank, R, Guy, H.R, Ruppersberg, J.P, Kalbitzer, H.R. | | Deposit date: | 1996-11-15 | | Release date: | 1997-06-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of inactivation gates from mammalian voltage-dependent potassium channels.
Nature, 385, 1997
|
|
1ZTN
 
 | | INACTIVATION GATE OF POTASSIUM CHANNEL RAW3, NMR, 8 STRUCTURES | | Descriptor: | Potassium voltage-gated channel subfamily C member 4 | | Authors: | Antz, C, Geyer, M, Fakler, B, Schott, M, Frank, R, Guy, H.R, Ruppersberg, J.P, Kalbitzer, H.R. | | Deposit date: | 1996-11-15 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of inactivation gates from mammalian voltage-dependent potassium channels.
Nature, 385, 1997
|
|
2B3A
 
 | | Solution structure of the Ras-binding domain of the Ral Guanosine Dissociation Stimulator | | Descriptor: | Ral guanine nucleotide dissociation stimulator | | Authors: | Gronwald, W, Maurer, T, Fuechsl, R, Wohlgemuth, S, Herrmann, C, Kalbitzer, H.R. | | Deposit date: | 2005-09-20 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | New insights into binding of the possible cancer target RalGDS
To be Published
|
|
2RGF
 
 | | RBD OF RAL GUANOSINE-NUCLEOTIDE EXCHANGE FACTOR (PROTEIN), NMR, 10 STRUCTURES | | Descriptor: | RALGEF-RBD | | Authors: | Geyer, M, Herrmann, C, Wittinghofer, A, Kalbitzer, H.R. | | Deposit date: | 1997-02-13 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ras-binding domain of RalGEF and implications for Ras binding and signalling.
Nat.Struct.Biol., 4, 1997
|
|
3L8Y
 
 | | Complex of Ras with cyclen | | Descriptor: | 1,4,7,10-tetraazacyclododecane, CALCIUM ION, GTPase HRas, ... | | Authors: | Rosnizeck, I.C, Graf, T, Spoerner, M, Traenkle, J, Filchtinski, D, Herrmann, C, Gremer, L, Vetter, I.R, Wittinghofer, A, Koenig, B, Kalbitzer, H.R. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Stabilizing a weak binding state for effectors in the human ras protein by cyclen complexes
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3L8Z
 
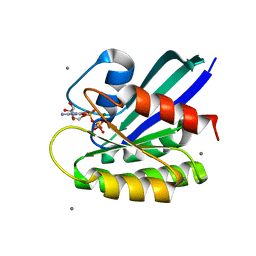 | | H-Ras wildtype new crystal form | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Rosnizeck, I.C, Graf, T, Spoerner, M, Traenkle, J, Filchtinski, D, Herrmann, C, Gremer, L, Vetter, I.R, Wittinghofer, A, Koenig, B, Kalbitzer, H.R. | | Deposit date: | 2010-01-04 | | Release date: | 2011-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Stabilizing a weak binding state for effectors in the human ras protein by cyclen complexes
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
8OSO
 
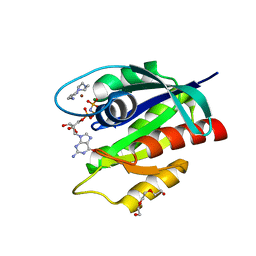 | | GTPase HRAS in complex with Zn-cyclen under 500 MPa pressure | | Descriptor: | 1,4,7,10-tetraazacyclododecane, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Colloc'h, N, Prange, T, Girard, E, Kalbitzer, H.R. | | Deposit date: | 2023-04-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Pressure Promotes Binding of the Allosteric Inhibitor Zn 2+ -Cyclen in Crystals of Activated H-Ras.
Chemistry, 30, 2024
|
|
8OSM
 
 | | GTPASE HRAS IN COMPLEX WITH ZN-CYCLEN AT 200 MPA PRESSURE | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Colloc'h, N, Girard, E, Prange, T, Kalbitzer, H.R. | | Deposit date: | 2023-04-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High Pressure Promotes Binding of the Allosteric Inhibitor Zn 2+ -Cyclen in Crystals of Activated H-Ras.
Chemistry, 30, 2024
|
|
8OSN
 
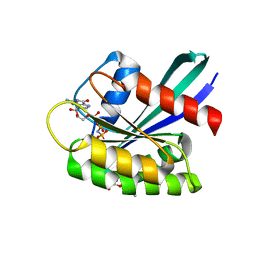 | | GTPASE HRAS IN COMPLEX WITH ZN-CYCLEN AT AMBIENT PRESSURE | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Colloc'h, N, Prange, T, Girard, E, Kalbitzer, H.R. | | Deposit date: | 2023-04-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Pressure Promotes Binding of the Allosteric Inhibitor Zn 2+ -Cyclen in Crystals of Activated H-Ras.
Chemistry, 30, 2024
|
|
1KN7
 
 | | Solution structure of the tandem inactivation domain (residues 1-75) of potassium channel RCK4 (Kv1.4) | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN KV1.4 | | Authors: | Wissmann, R, Bildl, W, Oliver, D, Beyermann, M, Kalbitzer, H.R, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-18 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Function of the "Tandem Inactivation Domain" of the Neuronal A-type
Potassium Channel Kv1.4
J.Biol.Chem., 278, 2003
|
|
1UB1
 
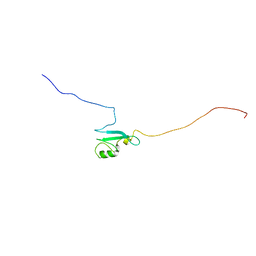 | | Solution structure of the matrix attachment region-binding domain of chicken MeCP2 | | Descriptor: | attachment region binding protein | | Authors: | Heitmann, B, Maurer, T, Weitzel, J.M, Stratling, W.H, Kalbitzer, H.R, Brunner, E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the matrix attachment region-binding domain of chicken MeCP2
EUR.J.BIOCHEM., 270, 2003
|
|
1IAQ
 
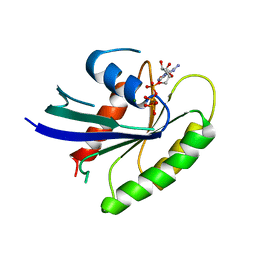 | | C-H-RAS P21 PROTEIN MUTANT WITH THR 35 REPLACED BY SER (T35S) COMPLEXED WITH GUANOSINE-5'-[B,G-IMIDO] TRIPHOSPHATE | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Spoerner, M, Herrmann, C, Vetter, I.R, Kalbitzer, H.R, Wittinghofer, A. | | Deposit date: | 2001-03-23 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic properties of the Ras switch I region and its importance for binding to effectors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G6P
 
 | | SOLUTION NMR STRUCTURE OF THE COLD SHOCK PROTEIN FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | COLD SHOCK PROTEIN TMCSP | | Authors: | Kremer, W, Schuler, B, Harrieder, S, Geyer, M, Gronwald, W, Welker, C, Jaenicke, R, Kalbitzer, H.R. | | Deposit date: | 2000-11-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur.J.Biochem., 268, 2001
|
|
1ALG
 
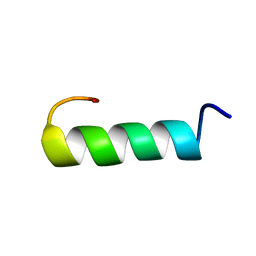 | | SOLUTION STRUCTURE OF AN HGR INHIBITOR, NMR, 10 STRUCTURES | | Descriptor: | P11 | | Authors: | Schott, M.K, Nordhoff, A, Becker, K, Kalbitzer, H.R, Schirmer, R.H. | | Deposit date: | 1997-06-03 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Denaturation and reactivation of dimeric human glutathione reductase--an assay for folding inhibitors.
Eur.J.Biochem., 245, 1997
|
|
1B4G
 
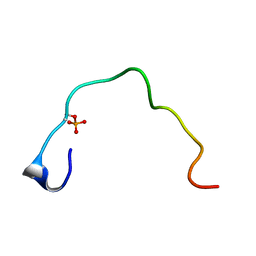 | | CONTROL OF K+ CHANNEL GATING BY PROTEIN PHOSPHORYLATION: STRUCTURAL SWITCHES OF THE INACTIVATION GATE, NMR, 22 STRUCTURES | | Descriptor: | POTASSIUM CHANNEL | | Authors: | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-04-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
1B4I
 
 | | Control of K+ Channel Gating by protein phosphorylation: structural switches of the inactivation gate, NMR, 22 structures | | Descriptor: | POTASSIUM CHANNEL | | Authors: | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-04-27 | | Last modified: | 2022-03-23 | | Method: | SOLUTION NMR | | Cite: | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
1QA5
 
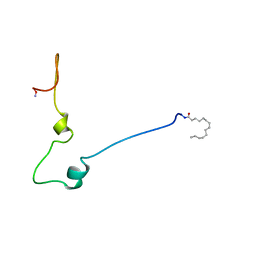 | |
