5IYT
 
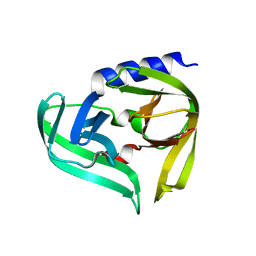 | | Complex structure of EV-B93 main protease 3C with N-Ethyl 4-((1-cycloheptyl-1,2-dihydropyrazol-3-one-5-yl)-amino)-4-oxo-2Z-butenamide | | Descriptor: | EV-B93 main protease 3C, N-Ethyl 4-((1-cycloheptyl-1,2-dihydropyrazol-3-one-5-yl)-amino)-4-oxo-butanamide | | Authors: | Kaczmarska, Z, Becker, D, Rademann, J, Coll, M. | | Deposit date: | 2016-03-24 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Irreversible inhibitors of the 3C protease of Coxsackie virus through templated assembly of protein-binding fragments.
Nat Commun, 7, 2016
|
|
7PIK
 
 | | Cryo-EM structure of E. coli TnsB in complex with right end fragment of Tn7 transposon | | Descriptor: | Right end fragment of Tn7 transposon, Transposon Tn7 transposition protein TnsB | | Authors: | Kaczmarska, Z, Czarnocki-Cieciura, M, Rawski, M, Nowotny, M. | | Deposit date: | 2021-08-20 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis of transposon end recognition explains central features of Tn7 transposition systems.
Mol.Cell, 82, 2022
|
|
5LKX
 
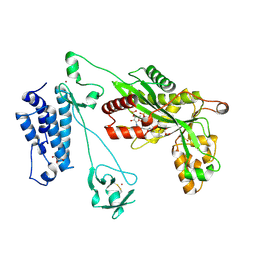 | | Crystal structure of the p300 acetyltransferase catalytic core with propionyl-coenzyme A. | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Histone acetyltransferase p300,Histone acetyltransferase p300, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
3RUO
 
 | | Complex structure of HevB EV93 main protease 3C with Rupintrivir (AG7088) | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, CHLORIDE ION, HEVB EV93 3C PROTEASE, ... | | Authors: | Kaczmarska, Z, Janowski, R, Costenaro, L, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
6H3R
 
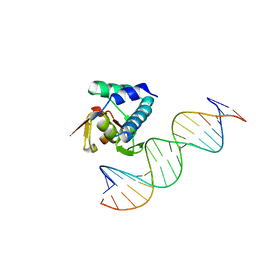 | |
6FZS
 
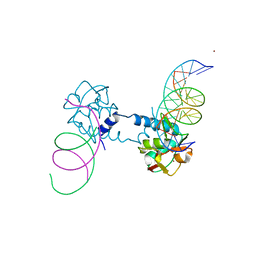 | | Crystal structure of Smad5-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 5, ZINC ION | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
6FZT
 
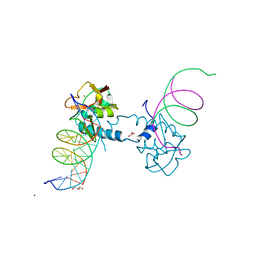 | | Crystal structure of Smad8_9-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), GLYCEROL, Mothers against decapentaplegic homolog 9, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
5LKU
 
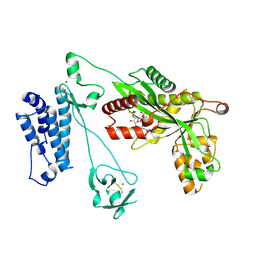 | | Crystal structure of the p300 acetyltransferase catalytic core with coenzyme A. | | Descriptor: | COENZYME A, Histone acetyltransferase p300,Histone acetyltransferase p300, ZINC ION | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5MEZ
 
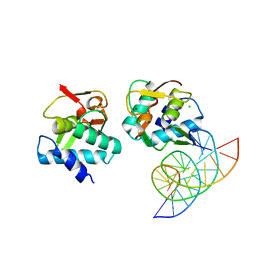 | | Crystal structure of Smad4-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*GP*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5LKT
 
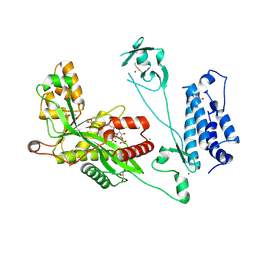 | | Crystal structure of the p300 acetyltransferase catalytic core with butyryl-coenzyme A. | | Descriptor: | Butyryl Coenzyme A, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-24 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5LKZ
 
 | | Crystal structure of the p300 acetyltransferase catalytic core with crotonyl-coenzyme A. | | Descriptor: | CROTONYL COENZYME A, GLYCEROL, Histone acetyltransferase p300,Histone acetyltransferase p300, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
5OD6
 
 | |
5MEY
 
 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MF0
 
 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5NM9
 
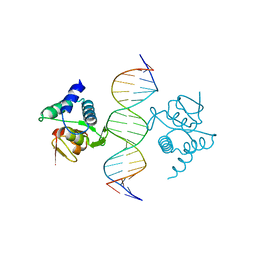 | | Crystal structure of the placozoa Trichoplax adhaerens Smad4-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*AP*TP*GP*CP*GP*GP*GP*CP*GP*CP*GP*CP*CP*CP*GP*CP*AP*T)-3'), Mothers against decapentaplegic homolog, ZINC ION | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5ODG
 
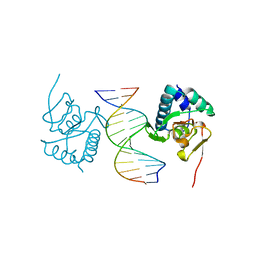 | | Crystal structure of Smad3-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 3, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
3Q3Y
 
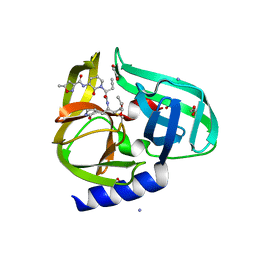 | | Complex structure of HEVB EV93 main protease 3C with Compound 1 (AG7404) | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, HEVB EV93 3C protease, ... | | Authors: | Costenaro, L, Kaczmarska, Z, Arnan, C, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
6EY7
 
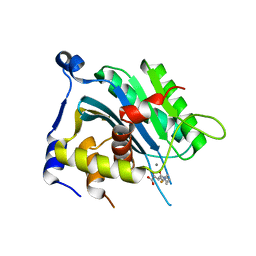 | | Human cytomegalovirus terminase nuclease domain, Mn soaked, inhibitor bound | | Descriptor: | 4-[(4-fluorophenyl)methyl-methyl-amino]-2,4-bis(oxidanylidene)butanoic acid, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Bongarzone, S, Nadal, M, Kaczmarska, Z, Machon, C, Alvarez, M, Albericio, F, Coll, M. | | Deposit date: | 2017-11-10 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Driven Discovery of alpha , gamma-Diketoacid Inhibitors Against UL89 Herpesvirus Terminase.
Acs Omega, 3, 2018
|
|
6ST8
 
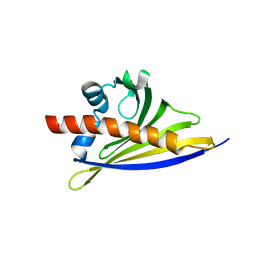 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
6STB
 
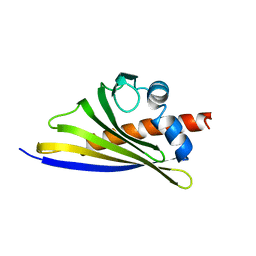 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, Q64W mutant | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
6STA
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, E46A D48A mutant | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
6ST9
 
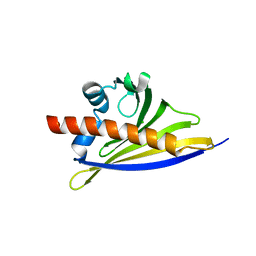 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, D48R mutant | | Descriptor: | CHLORIDE ION, Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
