2ARH
 
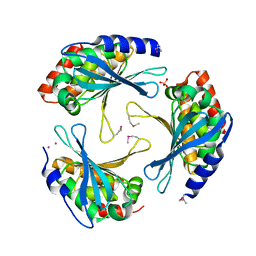 | | Crystal Structure of a Protein of Unknown Function AQ1966 from Aquifex aeolicus VF5 | | 分子名称: | CALCIUM ION, SELENIUM ATOM, SULFATE ION, ... | | 著者 | Qiu, Y, Kim, Y, Yang, X, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-08-19 | | 公開日 | 2005-10-04 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Crystal Structure of a Hypothetical Protein Aq_1966 from Aquifex aeolicus VF5
To be Published
|
|
6BIC
 
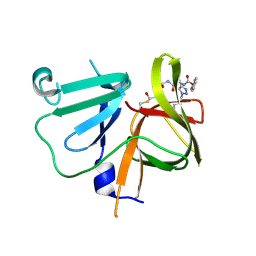 | | 2.25 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | 分子名称: | (phenylmethyl) ~{N}-[(9~{S},12~{S},15~{S})-9-(hydroxymethyl)-12-(2-methylpropyl)-6,11,14-tris(oxidanylidene)-1,5,10,13,18,19-hexazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate, 3C-like protease | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2017-11-01 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
6BIB
 
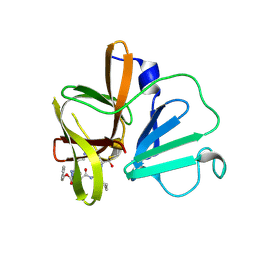 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | 分子名称: | 3C-like protease, benzyl [(9S,12S,15S)-12-(cyclohexylmethyl)-9-(hydroxymethyl)-6,11,14-trioxo-1,5,10,13,18,19-hexaazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2017-11-01 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
6BID
 
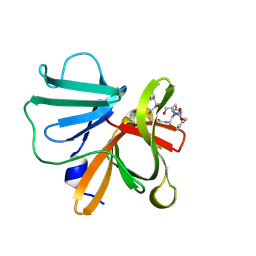 | | 1.15 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | 分子名称: | 3C-like protease, benzyl [(8S,11S,14S)-11-(cyclohexylmethyl)-8-(hydroxymethyl)-5,10,13-trioxo-1,4,9,12,17,18-hexaazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate | | 著者 | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2017-11-01 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
1CEK
 
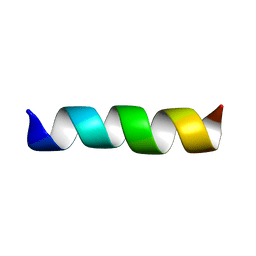 | | THREE-DIMENSIONAL STRUCTURE OF THE MEMBRANE-EMBEDDED M2 CHANNEL-LINING SEGMENT FROM THE NICOTINIC ACETYLCHOLINE RECEPTOR BY SOLID-STATE NMR SPECTROSCOPY | | 分子名称: | PROTEIN (ACETYLCHOLINE RECEPTOR M2) | | 著者 | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | 登録日 | 1999-03-09 | | 公開日 | 1999-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
6D35
 
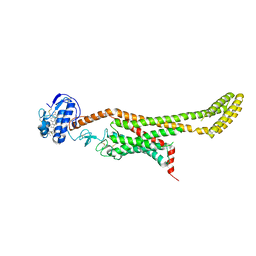 | | Crystal structure of Xenopus Smoothened in complex with cholesterol | | 分子名称: | CHOLESTEROL, Smoothened,Soluble cytochrome b562,Smoothened | | 著者 | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | 登録日 | 2018-04-14 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
2H7H
 
 | |
6D32
 
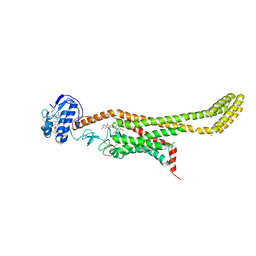 | | Crystal structure of Xenopus Smoothened in complex with cyclopamine | | 分子名称: | Cyclopamine, Smoothened,Soluble cytochrome b562,Smoothened | | 著者 | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | 登録日 | 2018-04-14 | | 公開日 | 2018-05-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.751 Å) | | 主引用文献 | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
1K9B
 
 | | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28 nm resolution. Structural peculiarities in a folded protein conformation | | 分子名称: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR | | 著者 | Voss, R.H, Ermler, U, Essen, L.O, Wenzl, G, Kim, Y.M, Flecker, P. | | 登録日 | 2001-10-29 | | 公開日 | 2001-11-16 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the bifunctional soybean Bowman-Birk inhibitor at 0.28-nm resolution. Structural peculiarities in a folded protein conformation.
Eur.J.Biochem., 242, 1996
|
|
1XNG
 
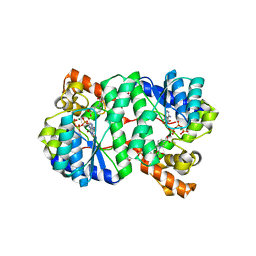 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | 著者 | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | 登録日 | 2004-10-05 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
1EQ8
 
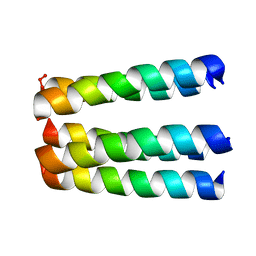 | | THREE-DIMENSIONAL STRUCTURE OF THE PENTAMERIC HELICAL BUNDLE OF THE ACETYLCHOLINE RECEPTOR M2 TRANSMEMBRANE SEGMENT | | 分子名称: | ACETYLCHOLINE RECEPTOR PROTEIN, HYDROXIDE ION | | 著者 | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | 登録日 | 2000-04-03 | | 公開日 | 2000-04-26 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
1XSI
 
 | | Structure of a Family 31 alpha glycosidase | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETIC ACID, Putative family 31 glucosidase yicI, ... | | 著者 | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | 登録日 | 2004-10-19 | | 公開日 | 2004-10-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanistic and Structural Analysis of a Family 31 alpha-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
7UH4
 
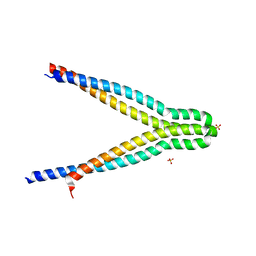 | | LXG-associated alpha-helical protein D2 (LapD2) | | 分子名称: | LXG-associated alpha-helical protein D2, SULFATE ION | | 著者 | Klein, T.A, Grebenc, D.W, Shah, P.Y, McArthur, O.D, Surette, M.G, Kim, Y, Whitney, J.C. | | 登録日 | 2022-03-25 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System.
Mbio, 13, 2022
|
|
3IQT
 
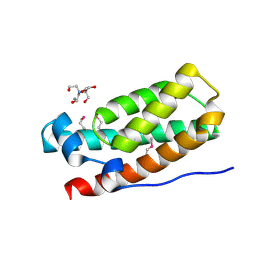 | | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073. | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Signal transduction histidine-protein kinase barA | | 著者 | Cuff, M.E, Rakowski, E, Kim, Y, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-08-20 | | 公開日 | 2009-09-22 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073.
TO BE PUBLISHED
|
|
3HLV
 
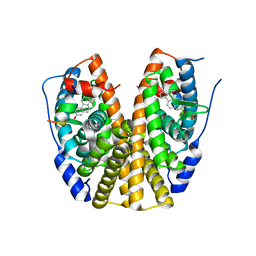 | | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and 16-alpha-hydroxy-estrone ((8S,9R,13S,14R,16R)-3,16-dihydroxy-13-methyl-7,8,9,11,12,14,15, 16-octahydro-6H-cyclopenta[a]phenanthren-17-one | | 分子名称: | (9beta,13alpha,16beta)-3,16-dihydroxyestra-1,3,5(10)-trien-17-one, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Rajan, S.S, Kim, Y, Vanek, K, Joachimiak, A, Greene, G.L. | | 登録日 | 2009-05-28 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of human alphaER-LBD in complex with GRIP peptide and 16alpha-hydroxyestrone
To be Published
|
|
3HM1
 
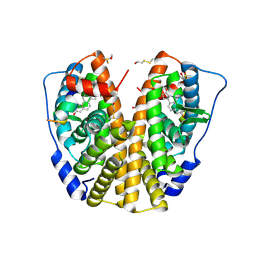 | | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15,16-octahydro-6H-cyclopenta[a]phenanthren-17-one) | | 分子名称: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Rajan, S.S, Kim, Y, Vanek, K, Liwanag, M, Joachimiak, A, Greene, G.L. | | 登録日 | 2009-05-28 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15, 16-octahydro-6H-cyclopenta[a]phenanthren-17-one)
To be Published
|
|
7F1W
 
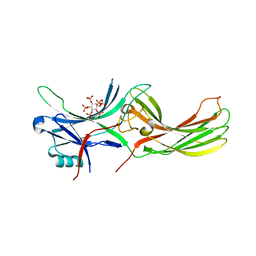 | | X-ray crystal structure of visual arrestin complexed with inositol hexaphosphate | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, S-arrestin | | 著者 | Kang, M, Jang, K, Eger, B.T, Ernst, O.P, Choe, H.W, Kim, Y.J. | | 登録日 | 2021-06-10 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.097 Å) | | 主引用文献 | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7F1X
 
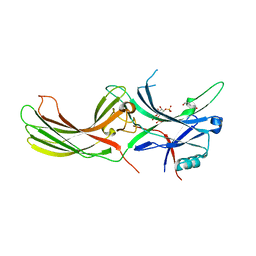 | | X-ray crystal structure of visual arrestin complexed with inositol 1,4,5-triphosphate | | 分子名称: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PENTANEDIAL, ... | | 著者 | Jang, K, Kang, M, Eger, B.T, Choe, H.W, Ernst, O.P, Kim, Y.J. | | 登録日 | 2021-06-10 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | 著者 | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | 登録日 | 2009-08-01 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (19.799999 Å) | | 主引用文献 | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1XSK
 
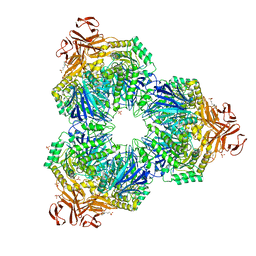 | | Structure of a Family 31 alpha glycosidase glycosyl-enzyme intermediate | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 5(R)-fluoro-beta-D-xylopyranose, Putative family 31 glucosidase yicI, ... | | 著者 | Lovering, A.L, Lee, S.S, Kim, Y.W, Withers, S.G, Strynadka, N.C. | | 登録日 | 2004-10-19 | | 公開日 | 2004-11-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanistic and Structural Analysis of a Family 31 {alpha}-Glycosidase and Its Glycosyl-enzyme Intermediate
J.Biol.Chem., 280, 2005
|
|
3F1I
 
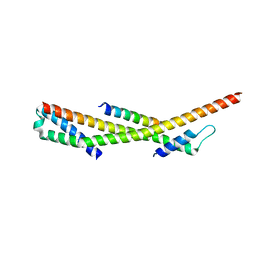 | | Human ESCRT-0 Core Complex | | 分子名称: | Hepatocyte growth factor-regulated tyrosine kinase substrate, Signal transducing adapter molecule 1 | | 著者 | Ren, X, Kloer, D.P, Kim, Y, Ghirlando, R, Saidi, L, Hummer, G, Hurley, J.H. | | 登録日 | 2008-10-28 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Hybrid Structural Model of the Complete Human ESCRT-0 Complex.
Structure, 17, 2009
|
|
5F3M
 
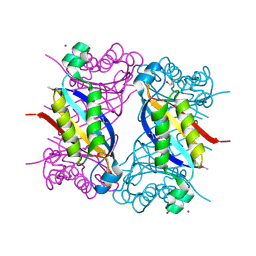 | | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complexed with L-neopterin at 1.5 Angstroms resolution . | | 分子名称: | 1,2-ETHANEDIOL, 7,8-dihydroneopterin aldolase, CHLORIDE ION, ... | | 著者 | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-12-03 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.498 Å) | | 主引用文献 | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complexed with L-neopterin at 1.5 Angstroms resolution .
To Be Published
|
|
2O5K
 
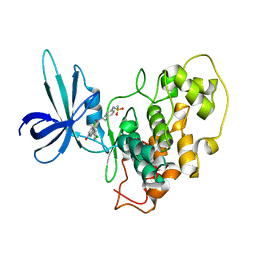 | | Crystal Structure of GSK3beta in complex with a benzoimidazol inhibitor | | 分子名称: | 2-(2,4-DICHLORO-PHENYL)-7-HYDROXY-1H-BENZOIMIDAZOLE-4-CARBOXYLIC ACID [2-(4-METHANESULFONYLAMINO-PHENYL)-ETHYL]-AMIDE, Glycogen synthase kinase-3 beta | | 著者 | Shin, D, Lee, S.C, Heo, Y.S, Cho, Y.S, Kim, Y.E, Hyun, Y.L, Cho, J.M, Lee, Y.S, Ro, S. | | 登録日 | 2006-12-06 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Design and synthesis of 7-hydroxy-1H-benzoimidazole derivatives as novel inhibitors of glycogen synthase kinase-3beta
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3ZGX
 
 | | Crystal structure of the kleisin-N SMC interface in prokaryotic condensin | | 分子名称: | CHROMOSOME PARTITION PROTEIN SMC, SEGREGATION AND CONDENSATION PROTEIN A | | 著者 | Burmann, F, Shin, H, Basquin, J, Soh, Y, Gimenez, V, Kim, Y, Oh, B, Gruber, S. | | 登録日 | 2012-12-19 | | 公開日 | 2013-01-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | An Asymmetric Smc-Kleisin Bridge in Prokaryotic Condensin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1OYI
 
 | | Solution structure of the Z-DNA binding domain of the vaccinia virus gene E3L | | 分子名称: | double-stranded RNA-binding protein | | 著者 | Kahmann, J.D, Wecking, D.A, Putter, V, Lowenhaupt, K, Kim, Y.-G, Schmieder, P, Oschkinat, H, Rich, A, Schade, M. | | 登録日 | 2003-04-04 | | 公開日 | 2004-03-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
