1M63
 
 | | Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes | | Descriptor: | CALCINEURIN B SUBUNIT ISOFORM 1, CALCIUM ION, CYCLOSPORIN A, ... | | Authors: | Huai, Q, Kim, H.-Y, Liu, Y, Zhao, Y, Mondragon, A, Liu, J.O, Ke, H. | | Deposit date: | 2002-07-12 | | Release date: | 2002-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Calcineurin-Cyclophilin-Cyclosporin Shows Common But Distinct Recognition of Immunophilin-Drug Complexes
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3TD4
 
 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with diaminopimelate | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
4ZZ7
 
 | | Crystal structure of methylmalonate-semialdehyde dehydrogenase (DddC) from Oceanimonas doudoroffii | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Do, H, Lee, C.W, Lee, S.G, Kang, H, Park, C.M, Kim, H.J, Park, H, Park, H, Lee, J.H. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii.
J. Microbiol., 54, 2016
|
|
3TD3
 
 | | Crystal structure of OmpA-like domain from Acinetobacter baumannii in complex with glycine | | Descriptor: | GLYCINE, Outer membrane protein omp38 | | Authors: | Park, J.S, Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane
Faseb J., 26, 2012
|
|
5XWB
 
 | | Crystal Structure of 5-Enolpyruvulshikimate-3-phosphate Synthase from a Psychrophilic Bacterium, Colwellia psychrerythraea | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Lee, J.H, Kim, H.J, Choi, J.M, Kim, D.-W, Seo, Y.-S. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 5-enolpyruvylshikimate-3-phosphate synthase from a psychrophilic bacterium, Colwellia psychrerythraea 34H.
Biochem. Biophys. Res. Commun., 492, 2017
|
|
2ZFC
 
 | | X-ray crystal structure of an engineered N-terminal HIV-1 GP41 trimer with enhanced stability and potency | | Descriptor: | HIV-1 GP41 | | Authors: | Dwyer, J.J, Wilson, K.L, Martin, K, Seedorff, J.E, Hasan, A, Kim, H. | | Deposit date: | 2007-12-29 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of an engineered N-terminal HIV-1 gp41 trimer with enhanced stability and potency
Protein Sci., 17, 2008
|
|
2HK1
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the presence of D-fructose | | Descriptor: | D-PSICOSE 3-EPIMERASE, D-fructose, MANGANESE (II) ION | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
2HK0
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the absence of substrate | | Descriptor: | D-PSICOSE 3-EPIMERASE | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
1DL4
 
 | | THE SOLUTION STRUCTURE OF A BAY-REGION 1S-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, DNA (5'-D(*CP*GP*GP*AP*CP*(BZA)AP*AP*GP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Li, Z, Kim, H.-Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-01-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (1S,2R,3S,4R)-N6-[1-(1,2,3,4-tetrahydro-2,3, 4-trihydroxybenz[a]anthracenyl)]-2'-deoxyadenosyl adduct in an oligodeoxynucleotide containing the human N-ras codon 61 sequence.
Biochemistry, 38, 1999
|
|
5IPW
 
 | | oligopeptide-binding protein OppA | | Descriptor: | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein, putative | | Authors: | Lee, H.H, Kim, H.J, Yoon, H.J. | | Deposit date: | 2016-03-10 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a putative oligopeptide-binding periplasmic protein from a hyperthermophile
Extremophiles, 20, 2016
|
|
1DJD
 
 | | THE SOLUTION STRUCTURE OF A NON-BAY REGION 11R-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 8,9,10,11-TETRAHYDRO-BENZO[A]ANTHRACENE-8,9,10-TRIOL, DNA(5'-D(*CT*GT*GT*AT*CT*AT*AT*GT*AT*AT*G)-3'), DNA(5'-D(*CT*TT*TT*CT*TT*TT*GT*TT*CT*CT*G)-3') | | Authors: | Li, Z, Kim, H.Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-12-02 | | Release date: | 1999-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Role of a polycyclic aromatic hydrocarbon bay region ring in modulating DNA adduct structure: the non-bay region (8S,9R,10S, 11R)-N(6)-[11-(8,9,10,11-tetrahydro-8,9, 10-trihydroxybenz[a]anthracenyl)]-2' -deoxyadenosyl adduct in codon 61 of the human N-ras protooncogene
Biochemistry, 38, 1999
|
|
2HJW
 
 | | Crystal Structure of the BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2006-07-02 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the biotin carboxylase domain of human acetyl-CoA carboxylase 2.
Proteins, 70, 2008
|
|
1Y4S
 
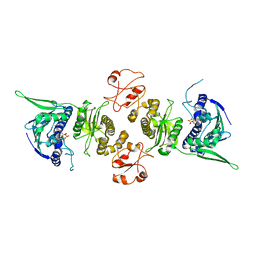 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, MAGNESIUM ION | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
3P28
 
 | |
1Y4U
 
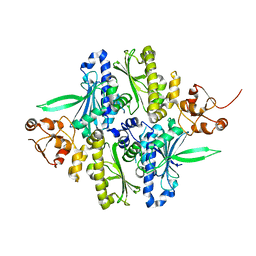 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
6P5W
 
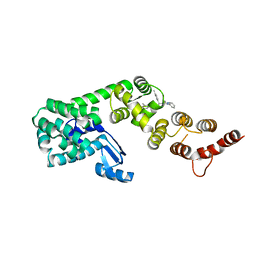 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6OI5
 
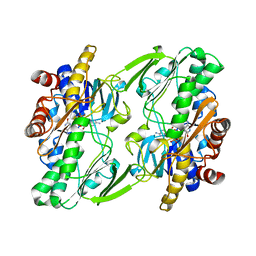 | | Crystal structure of human Sulfide Quinone Oxidoreductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Sulfide:quinone oxidoreductase, mitochondrial | | Authors: | Banerjee, R, Cho, U.S, Kim, H, Moon, S. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation.
Cell Chem Biol, 26, 2019
|
|
4OH8
 
 | | Crystal Structure of the human MST1-RASSF5 SARAH heterodimer | | Descriptor: | Ras association domain-containing protein 5, Serine/threonine-protein kinase 4 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6LT4
 
 | | AAA+ ATPase, ClpL from Streptococcus pneumoniae: ATPrS-bound | | Descriptor: | ATP-dependent Clp protease, ATP-binding subunit, MAGNESIUM ION, ... | | Authors: | Kim, G, Lee, S.G, Han, S, Jung, J, Jeong, H.S, Hyun, J.K, Rhee, D.K, Kim, H.M, Lee, S. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | ClpL is a functionally active tetradecameric AAA+ chaperone, distinct from hexameric/dodecameric ones.
Faseb J., 34, 2020
|
|
6P5V
 
 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
7N99
 
 | | SDE2 SAP domain apo structure | | Descriptor: | Isoform 2 of Replication stress response regulator SDE2 | | Authors: | Paung, Y, Weinheimer, A.S, Rageul, J, Khan, A, Ho, B, Tong, M, Alphonse, S, Seeliger, M.A, Kim, H. | | Deposit date: | 2021-06-17 | | Release date: | 2022-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Extended DNA-binding interfaces beyond the canonical SAP domain contribute to the function of replication stress regulator SDE2 at DNA replication forks.
J.Biol.Chem., 298, 2022
|
|
7F3G
 
 | |
5V7I
 
 | | Crystal structure of homo sapiens serine hydroxymethyltransferase 2 (mitochondrial) (SHMT2), in complex with glycine, PLP and folate-competitive pyrazolopyran inhibitor: 6-amino-4-isopropyl-3-methyl-4-(3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl)-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile | | Descriptor: | (4R)-6-amino-3-methyl-4-(propan-2-yl)-4-[3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl]-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ducker, G.S, Ghergurovich, J.M, Mainolfi, N, Suri, V, Jeong, S, Friedman, A, Manfredi, M, Kim, H, Rabinowitz, J.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4OH9
 
 | | Crystal Structure of the human MST2 SARAH homodimer | | Descriptor: | Serine/threonine-protein kinase 3 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NU2
 
 | | Crystal structure of an ice-binding protein (FfIBP) from the Antarctic bacterium, Flavobacterium frigoris PS1 | | Descriptor: | Antifreeze protein | | Authors: | Do, H, Kim, S.J, Lee, S.G, Park, H, Kim, H.J, Lee, J.H. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based characterization and antifreeze properties of a hyperactive ice-binding protein from the Antarctic bacterium Flavobacterium frigoris PS1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
