6BCE
 
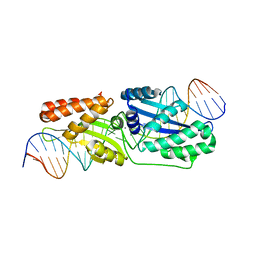 | | Wild-type I-LtrI bound to cognate substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (27-MER), Ribosomal protein 3/homing endonuclease-like fusion protein | | Authors: | Brown, C, Zhang, K, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site residue identity regulates cleavage preference of LAGLIDADG homing endonucleases.
Nucleic Acids Res., 46, 2018
|
|
2I5F
 
 | | Crystal structure of the C-terminal PH domain of pleckstrin in complex with D-myo-Ins(1,2,3,5,6)P5 | | Descriptor: | (1R,2R,3R,4R,5S,6S)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], Pleckstrin | | Authors: | Jackson, S.G, Haslam, R.J, Junop, M.S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1,2,3,5,6-pentakisphosphate.
Bmc Struct.Biol., 7, 2007
|
|
2I5C
 
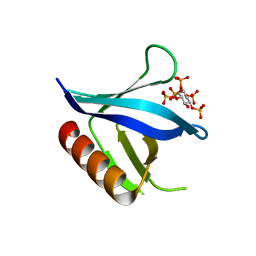 | | Crystal structure of the C-terminal PH domain of pleckstrin in complex with D-myo-Ins(1,2,3,4,5)P5 | | Descriptor: | (1R,2S,3R,4S,5S,6R)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], Pleckstrin | | Authors: | Jackson, S.G, Haslam, R.J, Junop, M.S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1,2,3,5,6-pentakisphosphate.
Bmc Struct.Biol., 7, 2007
|
|
2GQN
 
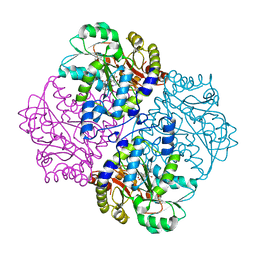 | |
2GMW
 
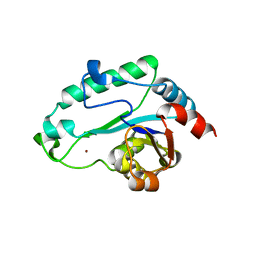 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli. | | Descriptor: | D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Zhang, K, DeLeon, G, Wright, G.D, Junop, M.S. | | Deposit date: | 2006-04-07 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and kinetic characterization of the LPS biosynthetic enzyme D-alpha,beta-D-heptose-1,7-bisphosphate phosphatase (GmhB) from Escherichia coli.
Biochemistry, 49, 2010
|
|
3EPU
 
 | | Crystal Structure of STM2138, a novel virulence chaperone in Salmonella | | Descriptor: | STM2138 Virulence Chaperone | | Authors: | Zhang, K, Andres, S.N, Hannemann, M, Coombes, B, Junop, M. | | Deposit date: | 2008-09-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis and quantitative proteomic
interactome of a novel virulence chaperone in Salmonella
To be Published
|
|
3L1V
 
 | | Crystal structure of GmhB from E. coli in complex with calcium and phosphate. | | Descriptor: | CALCIUM ION, D,D-heptose 1,7-bisphosphate phosphatase, PHOSPHATE ION, ... | | Authors: | Sugiman-Marangos, S.N, Junop, M.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and kinetic characterization of the LPS biosynthetic enzyme D-alpha,beta-D-heptose-1,7-bisphosphate phosphatase (GmhB) from Escherichia coli.
Biochemistry, 49, 2010
|
|
3ISS
 
 | | Crystal structure of enolpyruvyl-UDP-GlcNAc synthase (MurA):UDP-N-acetylmuramic acid:phosphite from Escherichia coli | | Descriptor: | PHOSPHITE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Jackson, S.G, Zhang, F, Chindemi, P, Junop, M.S, Berti, P.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence of Kinetic Control of Ligand Binding and Staged Product Release in MurA (Enolpyruvyl UDP-GlcNAc Synthase)-Catalyzed Reactions .
Biochemistry, 48, 2009
|
|
3L1U
 
 | | Crystal structure of Calcium-bound GmhB from E. coli. | | Descriptor: | CALCIUM ION, D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Sugiman-Marangos, S.N, Junop, M.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and kinetic characterization of the LPS biosynthetic enzyme D-alpha,beta-D-heptose-1,7-bisphosphate phosphatase (GmhB) from Escherichia coli.
Biochemistry, 49, 2010
|
|
4E8Y
 
 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | 7-O-phosphono-D-glycero-beta-D-manno-heptopyranose, CHLORIDE ION, D-beta-D-heptose 7-phosphate kinase, ... | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4E84
 
 | | Crystal Structure of Burkholderia cenocepacia HldA | | Descriptor: | 1,7-di-O-phosphono-D-glycero-beta-D-manno-heptopyranose, 7-O-phosphono-D-glycero-beta-D-manno-heptopyranose, CHLORIDE ION, ... | | Authors: | Lee, T.-W, Junop, M.S. | | Deposit date: | 2012-03-19 | | Release date: | 2012-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4EXW
 
 | |
3JYZ
 
 | | Crystal structure of Pseudomonas aeruginosa (strain: Pa110594) typeIV pilin in space group P41212 | | Descriptor: | SULFATE ION, Type IV pilin structural subunit | | Authors: | Nguyen, Y, Jackson, S.G, Aidoo, F, Junop, M.S, Burrows, L.L. | | Deposit date: | 2009-09-22 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural characterization of Novel Pseudomonas aeruginosa type IV pilins.
J.Mol.Biol., 395, 2010
|
|
3JZZ
 
 | | Crystal structure of Pseudomonas aeruginosa (strain: Pa110594) typeIV pilin in space group P212121 | | Descriptor: | Type IV pilin structural subunit | | Authors: | Nguyen, Y, Jackson, S.G, Aidoo, F, Junop, M.S, Burrows, L.L. | | Deposit date: | 2009-09-24 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Structural characterization of Novel Pseudomonas aeruginosa type IV pilins.
J.Mol.Biol., 395, 2010
|
|
4DRW
 
 | | Crystal Structure of the Ternary Complex between S100A10, an Annexin A2 N-terminal Peptide and an AHNAK Peptide | | Descriptor: | Neuroblast differentiation-associated protein AHNAK, Protein S100-A10/Annexin A2 chimeric protein | | Authors: | Rezvanpour, A, Lee, T.-W, Junop, M.S, Shaw, G.S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of an asymmetric ternary protein complex provides insight for membrane interaction.
Structure, 20, 2012
|
|
4E8W
 
 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | D-beta-D-heptose 7-phosphate kinase, POTASSIUM ION, {[2-({[5-(2,6-dimethoxyphenyl)-1,2,4-triazin-3-yl]amino}methyl)-1,3-benzothiazol-5-yl]oxy}acetic acid | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8654 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
4E8Z
 
 | | Crystal Structure of Burkholderia cenocepacia HldA in Complex with an ATP-competitive Inhibitor | | Descriptor: | D-beta-D-heptose 7-phosphate kinase, POTASSIUM ION, {[2-({[5-(2,6-dichlorophenyl)-1,2,4-triazin-3-yl]amino}methyl)-1,3-benzothiazol-5-yl]oxy}acetic acid | | Authors: | Lee, T.-W, Verhey, T.B, Junop, M.S. | | Deposit date: | 2012-03-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural-functional studies of Burkholderia cenocepacia D-glycero-beta-D-manno-heptose 7-phosphate kinase (HldA) and characterization of inhibitors with antibiotic adjuvant and antivirulence properties.
J.Med.Chem., 56, 2013
|
|
3II6
 
 | |
2I22
 
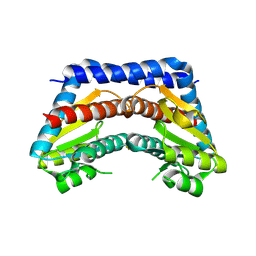 | | Crystal structure of Escherichia coli phosphoheptose isomerase in complex with reaction substrate sedoheptulose 7-phosphate | | Descriptor: | D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, Phosphoheptose isomerase | | Authors: | Blakely, K, Zhang, K, DeLeon, G, Wright, G, Junop, M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
2I2W
 
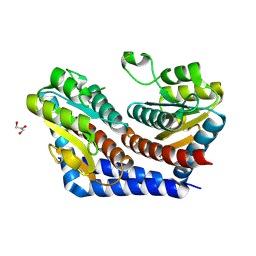 | | Crystal Structure of Escherichia Coli Phosphoheptose Isomerase | | Descriptor: | GLYCEROL, Phosphoheptose isomerase | | Authors: | DeLeon, G, Blakely, K, Zhang, K, Wright, G, Junop, M. | | Deposit date: | 2006-08-17 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
2HW2
 
 | |
4DZ1
 
 | |
4F6L
 
 | |
4F6C
 
 | |
