3GCB
 
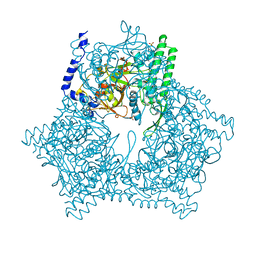 | | GAL6 (YEAST BLEOMYCIN HYDROLASE) MUTANT C73A/DELTAK454 | | Descriptor: | GAL6, GLYCEROL, SULFATE ION | | Authors: | Joshua-Tor, L, Zheng, W, Johnston, S.A. | | Deposit date: | 1998-02-27 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The unusual active site of Gal6/bleomycin hydrolase can act as a carboxypeptidase, aminopeptidase, and peptide ligase.
Cell(Cambridge,Mass.), 93, 1998
|
|
1A6R
 
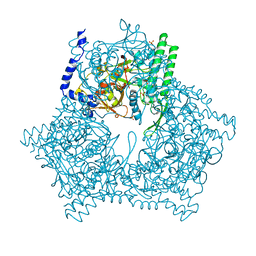 | | GAL6 (YEAST BLEOMYCIN HYDROLASE) MUTANT C73A | | Descriptor: | GAL6, SULFATE ION | | Authors: | Joshua-Tor, L, Zheng, W, Johnston, S.A. | | Deposit date: | 1998-02-27 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The unusual active site of Gal6/bleomycin hydrolase can act as a carboxypeptidase, aminopeptidase, and peptide ligase.
Cell(Cambridge,Mass.), 93, 1998
|
|
1D31
 
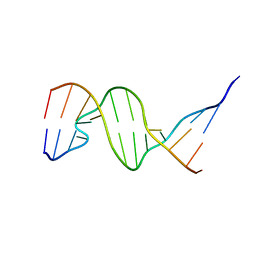 | | THE THREE-DIMENSIONAL STRUCTURES OF BULGE-CONTAINING DNA FRAGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Joshua-Tor, L, Frolow, F, Appella, E, Hope, H, Rabinovich, D, Sussman, J.L. | | Deposit date: | 1991-04-25 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures of bulge-containing DNA fragments.
J.Mol.Biol., 225, 1992
|
|
1GCB
 
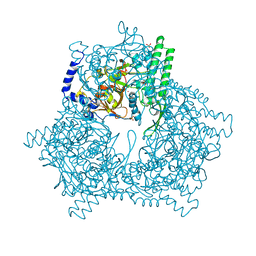 | | GAL6, YEAST BLEOMYCIN HYDROLASE DNA-BINDING PROTEASE (THIOL) | | Descriptor: | GAL6 HG (EMTS) DERIVATIVE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Joshua-Tor, L, Xu, H.E, Johnston, S.A, Rees, D.C. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6.
Science, 269, 1995
|
|
1R6Z
 
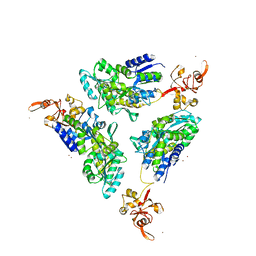 | | The Crystal Structure of the Argonaute2 PAZ domain (as a MBP fusion) | | Descriptor: | Chimera of Maltose-binding periplasmic protein and Argonaute 2, NICKEL (II) ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Song, J.J, Liu, J, Tolia, N.H, Schneiderman, J, Smith, S.K, Martienssen, R.A, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2003-10-17 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes.
Nat.Struct.Biol., 10, 2003
|
|
3BTV
 
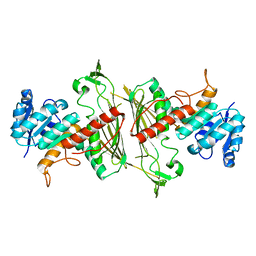 | |
7UX9
 
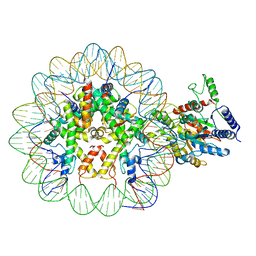 | | Arabidopsis DDM1 bound to nucleosome (H2A.W, H2B, H3.3, H4, with 147 bp DNA) | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Ipsaro, J.J, Adams, D.W, Joshua-Tor, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin remodeling of histone H3 variants by DDM1 underlies epigenetic inheritance of DNA methylation.
Cell, 186, 2023
|
|
3BTU
 
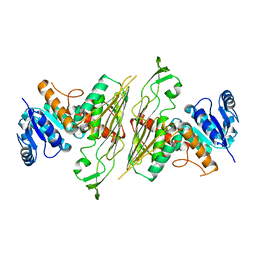 | |
1GSW
 
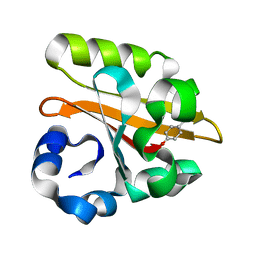 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSX
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G47S/G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSV
 
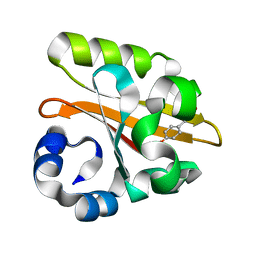 | | Crystal structure of the P65 crystal form of photoactive yellow protein G47S mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
6X46
 
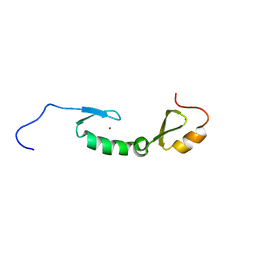 | | NMR solution structure of Asterix/Gtsf1 from mouse (CHHC zinc finger domains) | | Descriptor: | Gametocyte-specific factor 1, ZINC ION | | Authors: | Ipsaro, J.J, O'Brien, P.A, Bhattacharya, S, Palmer III, A.G, Joshua-Tor, L. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Asterix/Gtsf1 links tRNAs and piRNA silencing of retrotransposons.
Cell Rep, 34, 2021
|
|
2GXA
 
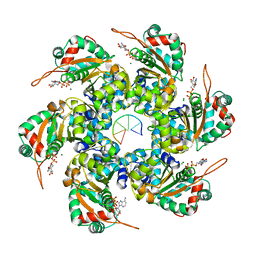 | |
9ASP
 
 | |
9ASN
 
 | |
9ASQ
 
 | |
9ASM
 
 | |
9ASO
 
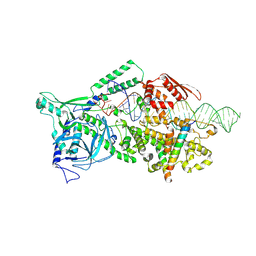 | |
6OU9
 
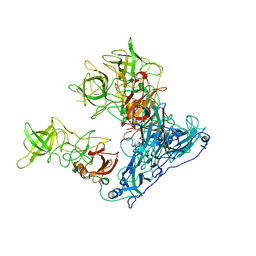 | | Asymmetric focused reconstruction of human norovirus GI.7 Houston strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Major capsid protein | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-04 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OUT
 
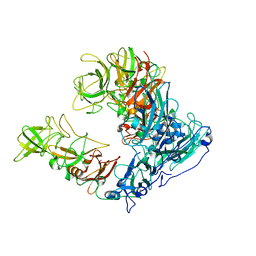 | | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Capsid protein VP1 | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8E29
 
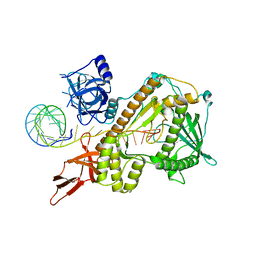 | |
8E27
 
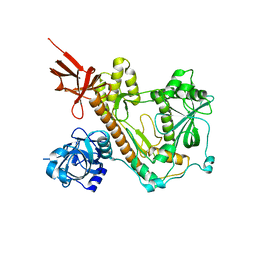 | | RNA-free Human Dis3L2 | | Descriptor: | DIS3-like exonuclease 2 | | Authors: | Meze, K, Thomas, D.R, Joshua-Tor, L. | | Deposit date: | 2022-08-14 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A shape-shifting nuclease unravels structured RNA.
Nat.Struct.Mol.Biol., 30, 2023
|
|
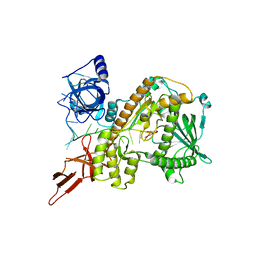
8E28
 
 | |
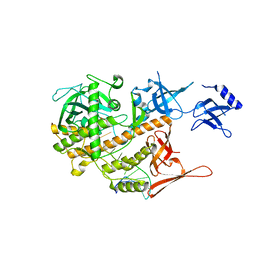
8E2A
 
 | |
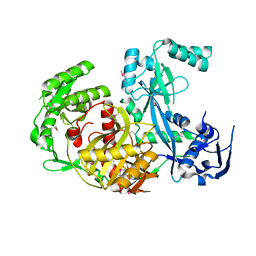
1U04
 
 | |
