7PFO
 
 | | Core human replisome | | Descriptor: | Cell division control protein 45 homolog, Claspin, DNA polymerase epsilon catalytic subunit A, ... | | Authors: | Jones, M.J, Yeeles, J.T.P. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a human replisome shows the organisation and interactions of a DNA replication machine.
Embo J., 40, 2021
|
|
7PLO
 
 | | H. sapiens replisome-CUL2/LRR1 complex | | Descriptor: | Cell division control protein 45 homolog, Claspin, Cullin-2, ... | | Authors: | Jones, M.J, Yeeles, J.T.P, Deegan, T.D, Jenkyn-Bedford, M. | | Deposit date: | 2021-09-01 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
6SXB
 
 | | XPF-ERCC1 Cryo-EM Structure, DNA-Bound form | | Descriptor: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*AP*TP*GP*CP*TP*GP*A)-3'), DNA excision repair protein ERCC-1, ... | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
6SXA
 
 | | XPF-ERCC1 Cryo-EM Structure, Apo-form | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
8B9D
 
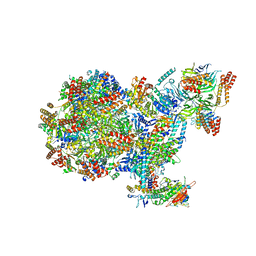 | | Human replisome bound by Pol Alpha | | Descriptor: | Cell division control protein 45 homolog, Claspin, DNA Molecule, ... | | Authors: | Jones, M.L, Yeeles, J.T.P. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | How Pol alpha-primase is targeted to replisomes to prime eukaryotic DNA replication.
Mol.Cell, 83, 2023
|
|
8B9C
 
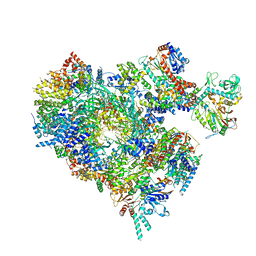 | | S. cerevisiae pol alpha - replisome complex | | Descriptor: | Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA polymerase alpha catalytic subunit A, ... | | Authors: | Jones, M.L, Yeeles, J.T.P. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | How Pol alpha-primase is targeted to replisomes to prime eukaryotic DNA replication.
Mol.Cell, 83, 2023
|
|
8B9B
 
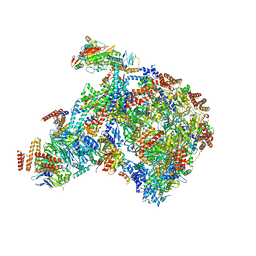 | |
8B9A
 
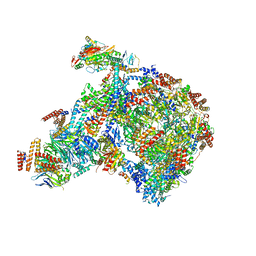 | |
2TGD
 
 | |
3V01
 
 | | Discovery of Novel Allosteric MEK Inhibitors Possessing Classical and Non-classical Bidentate Ser212 Interactions. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Heald, R, Jackson, P, Savy, P, Jones, M, Gancia, E, Burton, B, Newman, R, Boggs, J, Chan, E, Chan, J, Choo, E, Merchant, M, Ultsch, M, Wiesmann, C, Belvin, M, Price, S. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Discovery of Novel Allosteric Mitogen-Activated Protein Kinase Kinase (MEK) 1,2 Inhibitors Possessing Bidentate Ser212 Interactions.
J.Med.Chem., 55, 2012
|
|
1MPS
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH PHE M 197 REPLACED WITH ARG AND TYR M 177 REPLACED WITH PHE (CHAIN M, Y177F, F197R) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Mcauley-Hecht, K.E, Fyfe, P.K, Ridge, J.P, Prince, S, Hunter, C.N, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 1998-03-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural studies of wild-type and mutant reaction centers from an antenna-deficient strain of Rhodobacter sphaeroides: monitoring the optical properties of the complex from bacterial cell to crystal.
Biochemistry, 37, 1998
|
|
1W7E
 
 | |
1W7D
 
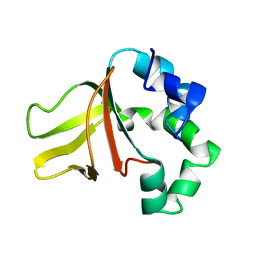 | |
6M9I
 
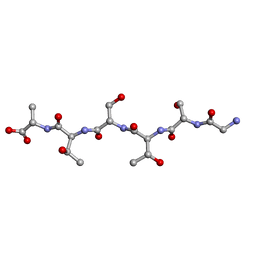 | | L-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | Ice nucleation protein | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6M7M
 
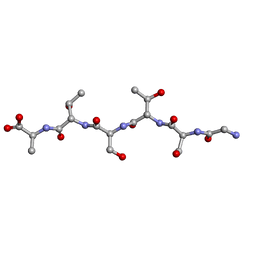 | | rac-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | L-GSTSTA from ice nucleation protein, inaZ, and its enantiomer, ... | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
6M9J
 
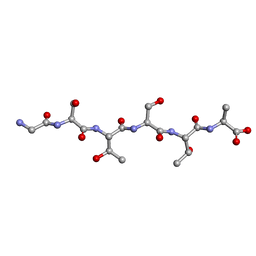 | | Racemic-GSTSTA from degenerate octameric repeats in InaZ, residues 707-712 | | Descriptor: | Ice nucleation protein | | Authors: | Zee, C, Glynn, C, Gallagher-Jones, M, Miao, J, Santiago, C.G, Cascio, D, Gonen, T, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Homochiral and racemic MicroED structures of a peptide repeat from the ice-nucleation protein InaZ.
IUCrJ, 6, 2019
|
|
7N2G
 
 | | MicroED structure of human CPEB3 segment(154-161) kinked polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.201 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2J
 
 | | MicroED structure of a mutant mammalian prion segment phased by ARCIMBOLDO-BORGES | | Descriptor: | prion protein | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2F
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2K
 
 | | MicroED structure of sequence variant of repeat segment of the yeast prion New1p phased by ARCIMBOLDO-BORGES | | Descriptor: | prion New1p | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.301 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2L
 
 | | MicroED structure of a mutant mammalian prion segment | | Descriptor: | prion protein | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.9 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2I
 
 | | MicroED structure of human LECT2 (45-53) phased by ARCIMBOLDO-BORGES | | Descriptor: | LECT2 | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.402 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2H
 
 | | X-Ray structure of a sequence variant of a repeat segment of the yeast prion New1p | | Descriptor: | prion New1p | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.102 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2D
 
 | | MicroED structure of human zinc finger protein 292 segment (534-542) phased by ARCIMBOLDO-BORGES | | Descriptor: | DIMETHYL SULFOXIDE, zinc finger protein 292 | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.503 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2E
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
