7RXS
 
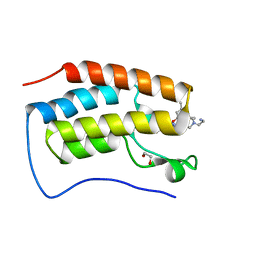 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXT
 
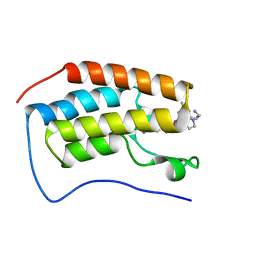 | | Crystal of BRD4(D1) with 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
1B35
 
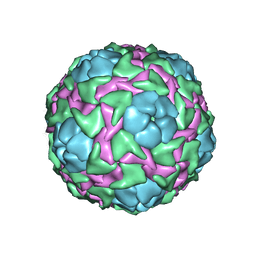 | | CRICKET PARALYSIS VIRUS (CRPV) | | Descriptor: | PROTEIN (CRICKET PARALYSIS VIRUS, VP1), VP2), ... | | Authors: | Tate, J.G, Liljas, L, Scotti, P.D, Christian, P.D, Lin, T.W, Johnson, J.E. | | Deposit date: | 1998-12-17 | | Release date: | 1999-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cricket paralysis virus: the first view of a new virus family.
Nat.Struct.Biol., 6, 1999
|
|
7MLQ
 
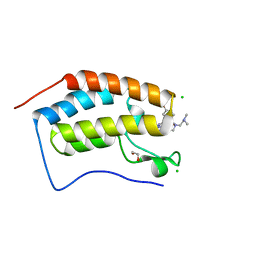 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (compound 26) | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Johnson, J.A, Vail, N.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7MLS
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine, Bromodomain-containing protein 4, ... | | Authors: | Cui, H, Johnson, J.A, Zahid, H, Buchholz, C.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-04-28 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
4NKM
 
 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
4NKD
 
 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
4NKO
 
 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.496 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
6XPG
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography | | Descriptor: | Lysophospholipase L1 | | Authors: | Kim, Y, Sherrell, D.A, Owen, R, Axford, D, Ebrahim, A, Johnson, J, Welk, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus by Serial Crystallography
To Be Published
|
|
6XPM
 
 | | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature | | Descriptor: | Lysophospholipase L1, SODIUM ION | | Authors: | Kim, Y, Johnson, J, Welk, L, Endres, M, Levens, A, Sherrell, D.A, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Sialate O-acetylesterase from Bacteroides vulgatus with microfluidics crystals at room temperature
To Be Published
|
|
3F2E
 
 | | Crystal structure of Yellowstone SIRV coat protein C-terminus | | Descriptor: | CITRIC ACID, SIRV coat protein | | Authors: | Taurog, R.E, Szymczyna, B.R, Williamson, J.R, Johnson, J.E. | | Deposit date: | 2008-10-29 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Synergy of NMR, computation, and X-ray crystallography for structural biology.
Structure, 17, 2009
|
|
1IF0
 
 | | PSEUDO-ATOMIC MODEL OF BACTERIOPHAGE HK97 PROCAPSID (PROHEAD II) | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN GP5) | | Authors: | Conway, J.F, Wikoff, W.R, Cheng, N, Duda, R.L, Hendrix, R.W, Johnson, J.E, Steven, A.C. | | Deposit date: | 2001-04-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Virus maturation involving large subunit rotations and local refolding.
Science, 292, 2001
|
|
3R0R
 
 | |
3S6P
 
 | |
7K3M
 
 | | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography | | Descriptor: | Beta-lactamase | | Authors: | Kim, Y, Sherrell, D.A, Johnson, J, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-11 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography
To Be Published
|
|
3DDX
 
 | | HK97 bacteriophage capsid Expansion Intermediate-II model | | Descriptor: | Major capsid protein | | Authors: | Lee, K.K, Gan, L, Conway, J.F, Hendrix, R.W, Steven, A.C, Johnson, J.E. | | Deposit date: | 2008-06-06 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY | | Cite: | Virus capsid expansion driven by the capture of mobile surface loops.
Structure, 16, 2008
|
|
5FMO
 
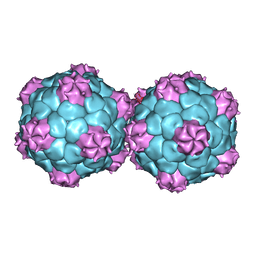 | | Crystal structure and proteomics analysis of empty virus like particles of Cowpea mosaic virus | | Descriptor: | EMPTY VIRUS LIKE PARTICLES OF COWPEA MOSAIC VIRUS | | Authors: | Huynh, N, Hesketh, E.L, Saxena, P, Meshcheriakova, Y, Ku, Y.C, Hoang, L, Johnson, J.E, Ranson, N.A, Lomonossoff, G.P, Reddy, V.S. | | Deposit date: | 2015-11-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure and Proteomics Analysis of Empty Virus-Like Particles of Cowpea Mosaic Virus.
Structure, 24, 2016
|
|
3E8K
 
 | |
1BMV
 
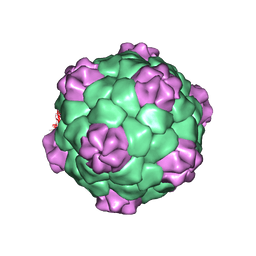 | | PROTEIN-RNA INTERACTIONS IN AN ICOSAHEDRAL VIRUS AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (ICOSAHEDRAL VIRUS - A DOMAIN), PROTEIN (ICOSAHEDRAL VIRUS - B AND C DOMAIN), RNA (5'-R(*GP*GP*UP*CP*AP*AP*AP*AP*UP*GP*C)-3') | | Authors: | Chen, Z, Stauffacher, C, Li, Y, Schmidt, T, Bomu, W, Kamer, G, Shanks, M, Lomonossoff, G, Johnson, J.E. | | Deposit date: | 1989-10-09 | | Release date: | 1989-10-09 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein-RNA interactions in an icosahedral virus at 3.0 A resolution.
Science, 245, 1989
|
|
3QPR
 
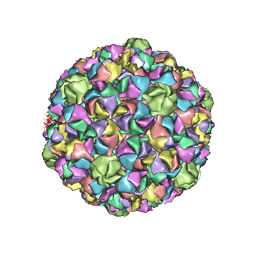 | | HK97 Prohead I encapsidating inactive virally encoded protease | | Descriptor: | Major capsid protein | | Authors: | Huang, R.K, Khayat, R, Lee, K.K, Gertsman, I, Duda, R.L, Hendrix, R.W, Johnson, J.E. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation.
J.Mol.Biol., 408, 2011
|
|
3BM9
 
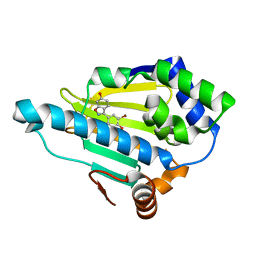 | | Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 | | Descriptor: | 4-bromo-6-(6-hydroxy-1,2-benzisoxazol-3-yl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gopalsamy, A, Shi, M, Vogan, E.M, Golas, J, Jacob, J, Johnson, J, Lee, F, Nilakantan, R, Peterson, R, Svenson, K, Tam, M.S, Wen, Y, Chopra, R, Ellingboe, J, Arndt, K, Boschelli, F. | | Deposit date: | 2007-12-12 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90.
J.Med.Chem., 51, 2008
|
|
7KW7
 
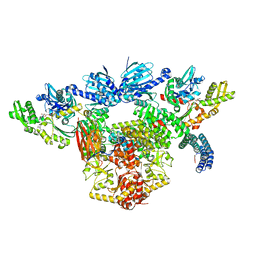 | | Atomic cryoEM structure of Hsp90-Hsp70-Hop-GR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glucocorticoid receptor, Heat shock 70 kDa protein 1A, ... | | Authors: | Wang, R.Y, Noddings, C.M, Kirschke, E, Myasnikov, A, Johnson, J.L, Agard, D.A. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism.
Nature, 601, 2022
|
|
3J31
 
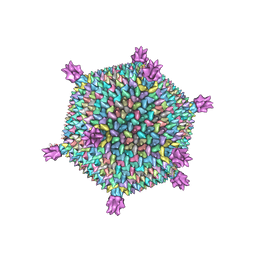 | | Life in the extremes: atomic structure of Sulfolobus Turreted Icosahedral Virus | | Descriptor: | A223 penton base, A55 membrane protein, C381 turret protein, ... | | Authors: | Veesler, D, Ng, T.S, Sendamarai, A.K, Eilers, B.J, Lawrence, C.M, Lok, S.M, Young, M.J, Johnson, J.E, Fu, C.-Y. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3BMY
 
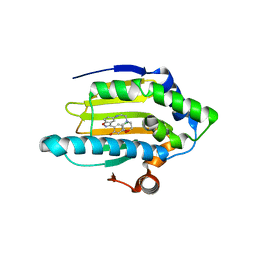 | | Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 | | Descriptor: | 4-chloro-6-{5-[(2-morpholin-4-ylethyl)amino]-1,2-benzisoxazol-3-yl}benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gopalsamy, A, Shi, M, Vogan, E.M, Golas, J, Jacob, J, Johnson, J, Lee, F, Nilakantan, R, Peterson, R, Svenson, K, Tam, M.S, Wen, Y, Chopra, R, Ellingboe, J, Arndt, K, Boschelli, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90.
J.Med.Chem., 51, 2008
|
|
7RSK
 
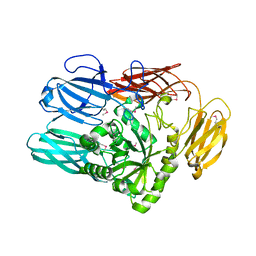 | | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus | | Descriptor: | Glycosyl hydrolase family 2, sugar binding domain protein | | Authors: | Kim, Y, Nocek, B, Endres, M, Joachimiak, G, Johnson, J, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-08-11 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus
To Be Published
|
|
