3IBV
 
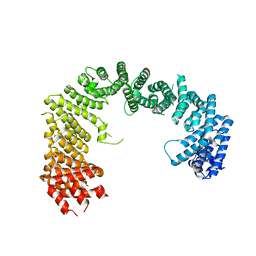 | | Karyopherin cytosolic state | | Descriptor: | CALCIUM ION, Exportin-T | | Authors: | Cook, A.G, Fukuhara, N, Jinek, M, Conti, E. | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the tRNA export factor in the nuclear and cytosolic states
Nature, 461, 2009
|
|
7Z4D
 
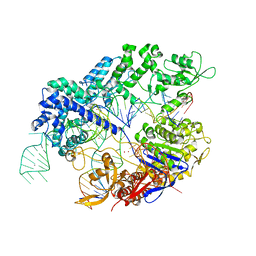 | |
7ZYH
 
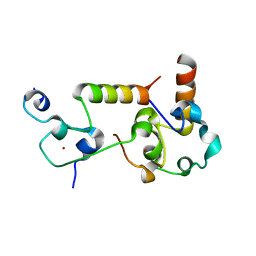 | | Crystal structure of human CPSF30 in complex with hFip1 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 4, Isoform 4 of Pre-mRNA 3'-end-processing factor FIP1, ZINC ION | | Authors: | Muckenfuss, L.M, Jinek, M, Migenda Herranz, A.C, Clerici, M. | | Deposit date: | 2022-05-24 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fip1 is a multivalent interaction scaffold for processing factors in human mRNA 3' end biogenesis.
Elife, 11, 2022
|
|
7ZY4
 
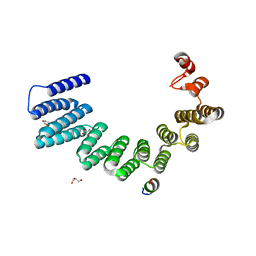 | |
7Z4E
 
 | | SpCas9 bound to 8-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 8 nucleotide complementary DNA substrate, Target strand of 8 nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4I
 
 | | SpCas9 bound to 16-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 16-nucleotide complementary DNA substrate, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4H
 
 | | SpCas9 bound to 14-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 14-nucleotide complementary DNA substrate, Target strand of 14-nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4C
 
 | | SpCas9 bound to 6 nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 6 nucleotide complementary DNA substrate, Target strand of 6 nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4G
 
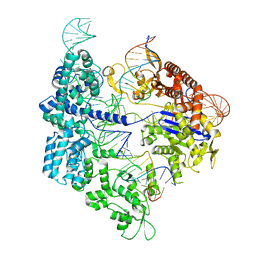 | | SpCas9 bound to 12-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 12-nucleotide complementary DNA substrate, Target strand of 12-nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4K
 
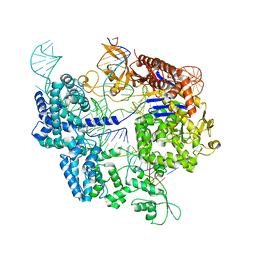 | | SpCas9 bound to 10-nucleotide complementary DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 10-nucleotide complementary DNA substrate, Target strand of 10-nucleotide complementary DNA substrate, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4J
 
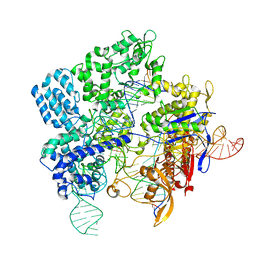 | |
7Z4L
 
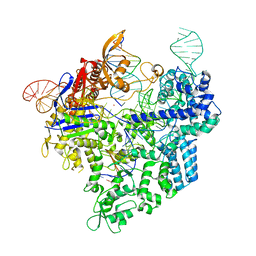 | |
6FUW
 
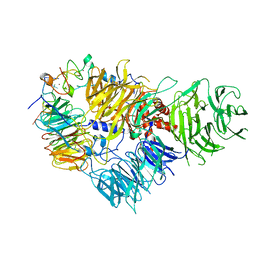 | | Cryo-EM structure of the human CPSF160-WDR33-CPSF30 complex bound to the PAS AAUAAA motif at 3.1 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*AP*GP*G)-3'), ... | | Authors: | Clerici, M, Faini, M, Jinek, M. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7ZO1
 
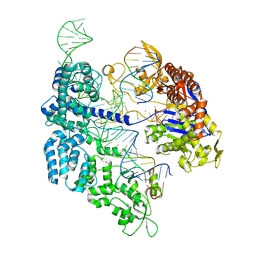 | | SpCas9 bound to CD34 off-target9 DNA substrate | | Descriptor: | CD34 off-target9 DNA non-target strand, CD34 off-target9 DNA target strand, CD34 sgRNA, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
6F9W
 
 | |
6F9N
 
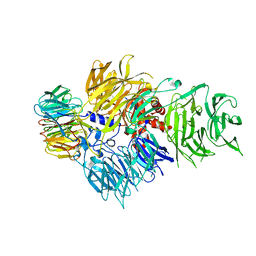 | | CRYSTAL STRUCTURE OF THE HUMAN CPSF160-WDR33 COMPLEX | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, pre-mRNA 3' end processing protein WDR33 | | Authors: | Clerici, M, Jinek, M. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex.
Elife, 6, 2017
|
|
5FQ5
 
 | | Crystal structure of Cas9-sgRNA-DNA complex solved by native SAD phasing | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Olieric, V, Weinert, T, Finke, A, Anders, C, Jinek, M, Wang, M. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Data-Collection Strategy for Challenging Native Sad Phasing.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5FW1
 
 | | Crystal structure of SpyCas9 variant VQR bound to sgRNA and TGAG PAM target DNA | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Bargsten, K, Jinek, M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
5FW2
 
 | | Crystal structure of SpCas9 variant EQR bound to sgRNA and TGAG PAM target DNA | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Bargsten, K, Jinek, M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.676 Å) | | Cite: | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
5FW3
 
 | | Crystal structure of SpCas9 variant VRER bound to sgRNA and TGCG PAM target DNA | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Bargsten, K, Jinek, M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
5FSH
 
 | |
3P3I
 
 | | Crystal structure of the F36A mutant of the fluoroacetyl-CoA-specific thioesterase FlK in complex with fluoroacetate and CoA | | Descriptor: | COENZYME A, Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P2R
 
 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase FlK in complex with fluoroacetate | | Descriptor: | Fluoroacetyl coenzyme A thioesterase, fluoroacetic acid | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-03 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P2S
 
 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase FlK in an open conformation | | Descriptor: | Fluoroacetyl coenzyme A thioesterase | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-04 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
3P2Q
 
 | | Crystal structure of the fluoroacetyl-CoA-specific thioesterase, FlK | | Descriptor: | Fluoroacetyl coenzyme A thioesterase | | Authors: | Weeks, A.M, Coyle, S.M, Jinek, M, Doudna, J.A, Chang, M.C.Y. | | Deposit date: | 2010-10-03 | | Release date: | 2010-10-20 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity.
Biochemistry, 49, 2010
|
|
