8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8WJL
 
 | | Cryo-EM structure of 6-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.15 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | Descriptor: | RNA polymerase-associated protein RapA, SULFATE ION | | Authors: | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
7Y5X
 
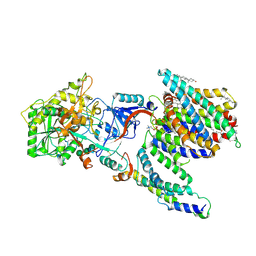 | | CryoEM structure of PS2-containing gamma-secretase treated with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5T
 
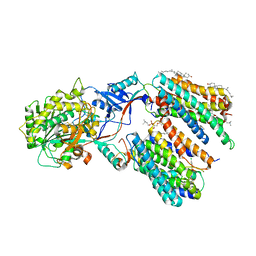 | | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7Y5Z
 
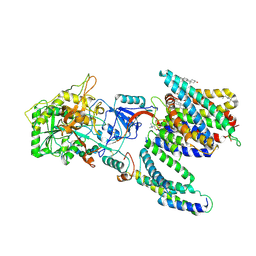 | | CryoEM structure of human PS2-containing gamma-secretase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-18 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
1YY7
 
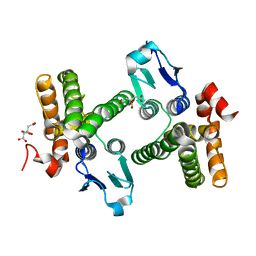 | | Crystal structure of stringent starvation protein A (SspA), an RNA polymerase-associated transcription factor | | Descriptor: | CITRIC ACID, stringent starvation protein A | | Authors: | Hansen, A.-M, Gu, Y, Li, M, Andrykovitch, M, Waugh, D.S, Jin, D.J, Ji, X. | | Deposit date: | 2005-02-23 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the function of stringent starvation protein A as a transcription factor
J.Biol.Chem., 280, 2005
|
|
2H80
 
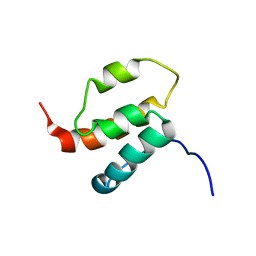 | | NMR structures of SAM domain of Deleted in Liver Cancer 2 (DLC2) | | Descriptor: | StAR-related lipid transfer protein 13 | | Authors: | Li, H.Y, Fung, K.L, Jin, D.Y, Chung, S.S, Ko, B.C, Sun, H.Z. | | Deposit date: | 2006-06-06 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2
Proteins, 67, 2007
|
|
4MQL
 
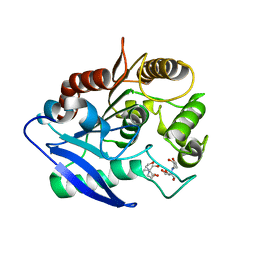 | | Crystal structure of Antigen 85C-C209S mutant | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
4QEK
 
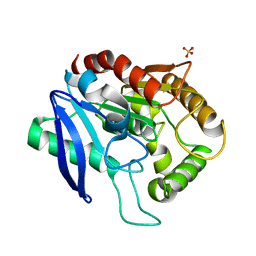 | | Crystal structure of Antigen 85C-S124A mutant | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, SULFATE ION, alpha-D-glucopyranose | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4MQM
 
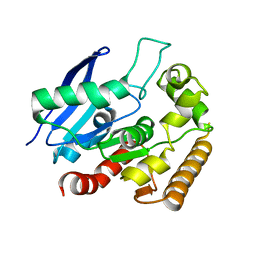 | | Crystal structure of Antigen 85C in presence of Ebselen | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
4QE3
 
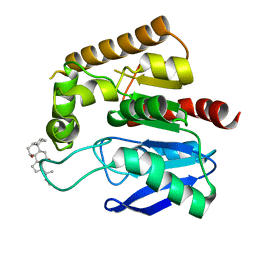 | | Crystal structure of Antigen 85C-H260Q mutant | | Descriptor: | ACETIC ACID, CHAPSO, Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QDT
 
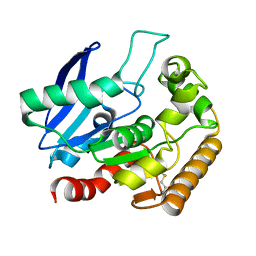 | |
4QDZ
 
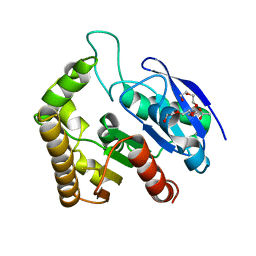 | | Crystal structure of Antigen 85C-E228Q mutant | | Descriptor: | ACETATE ION, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, HEXAETHYLENE GLYCOL | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QDX
 
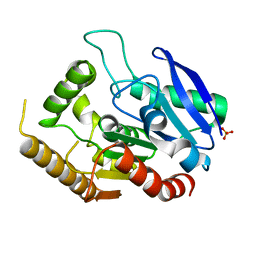 | | Crystal structure of Antigen 85C-C209G mutant | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, SULFATE ION | | Authors: | Favrot, L, Lajiness, D.H, Ronning, D.R. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Inactivation of the Mycobacterium tuberculosis Antigen 85 Complex by Covalent, Allosteric Inhibitors.
J.Biol.Chem., 289, 2014
|
|
4QDO
 
 | |
4QDU
 
 | |
5Z5R
 
 | | Nukacin ISK-1 in inactive state | | Descriptor: | Lantibiotic nukacin | | Authors: | Kohda, D, Fujinami, D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
5Z5Q
 
 | | Nukacin ISK-1 in active state | | Descriptor: | Lantibiotic nukacin | | Authors: | Kohda, D, Fujinami, D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
8JHE
 
 | | Hyper-thermostable ancestral L-amino acid oxidase 2 (HTAncLAAO2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Hyper thermostable ancestral L-amino acid oxidase | | Authors: | Kawamura, Y, Ishida, C, Miyata, R, Miyata, A, Hayashi, S, Fujinami, D, Ito, S, Nakano, S. | | Deposit date: | 2023-05-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional analysis of hyper-thermostable ancestral L-amino acid oxidase that can convert Trp derivatives to D-forms by chemoenzymatic reaction.
Commun Chem, 6, 2023
|
|
7D7E
 
 | | Structure of PKD1L3-CTD/PKD2L1 in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Polycystic kidney disease 2-like 1 protein, ... | | Authors: | Su, Q, Chen, M, Li, B, Wang, Y, Jing, D, Zhan, X, Yu, Y, Shi, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for Ca 2+ activation of the heteromeric PKD1L3/PKD2L1 channel.
Nat Commun, 12, 2021
|
|
