7N0H
 
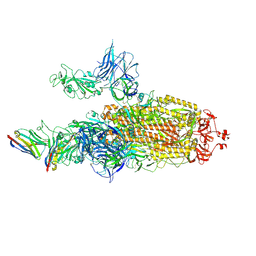 | | CryoEM structure of SARS-CoV-2 spike protein (S-6P, 2-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7N0G
 
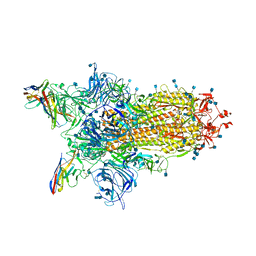 | | CryoEm structure of SARS-CoV-2 spike protein (S-6P, 1-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
5WCU
 
 | |
8I0C
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S0703 | | Descriptor: | 1-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]cyclopropane-1-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, He, S, Liu, Y, Fang, P, Sun, H. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
8TQ9
 
 | | Crystal structure of Fab.S19.8 in complex with MHC-I (H2-Dd) | | Descriptor: | Beta-2-microglobulin, Fab.S19.8 Heavy Chain, Fab.S19.8 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQA
 
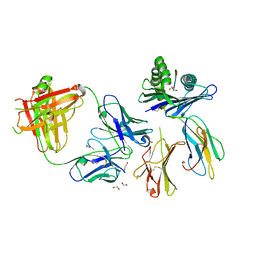 | | Crystal structure of Fab.28.14.8 in complex with MHC-I (H2-Db) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ7
 
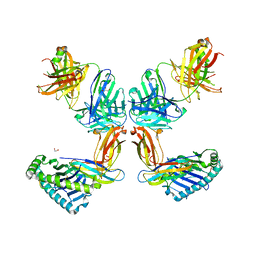 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
8TQ8
 
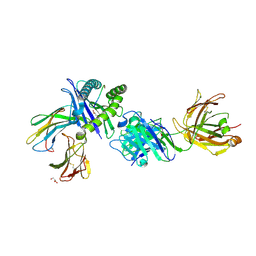 | | Crystal structure of Fab.34.5.8 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab.34.5.8 Heavy chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
3ULY
 
 | |
3UM3
 
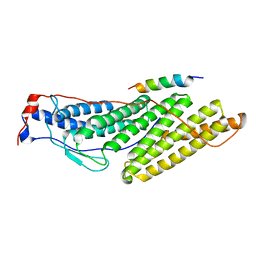 | |
3UM1
 
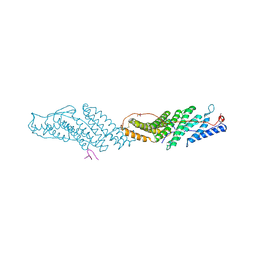 | |
3UM0
 
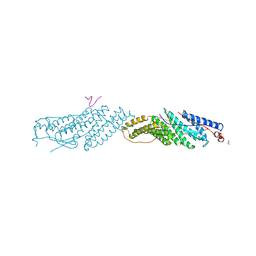 | |
3UM2
 
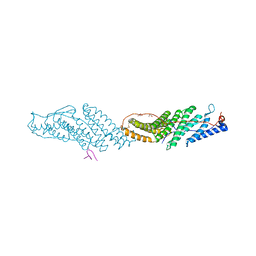 | |
3BM0
 
 | |
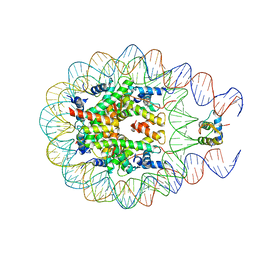
4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | Descriptor: | CITRIC ACID, DNA (167-mer), H5, ... | | Authors: | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
7TUH
 
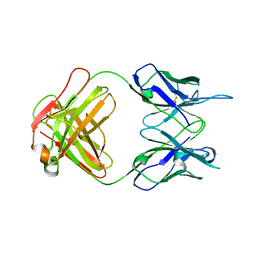 | | Crystal structure of anti-tapasin PaSta2-Fab | | Descriptor: | PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUD
 
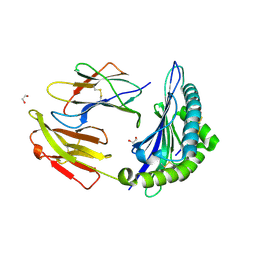 | | Crystal structure of HLA-B*44:05 (T73C) with 6mer EEFGRC and dipeptide GL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, EEFGRC peptide, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUG
 
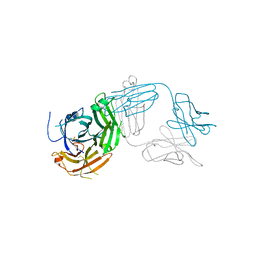 | | Crystal structure of Tapasin in complex with PaSta2-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUE
 
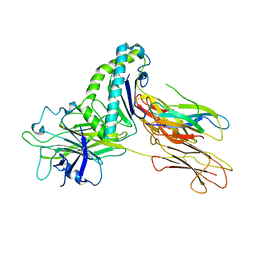 | | Crystal structure of Tapasin in complex with HLA-B*44:05 (T73C) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B alpha chain, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUF
 
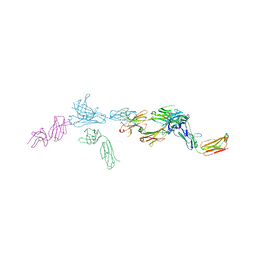 | | Crystal structure of Tapasin in complex with PaSta1-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta1 Fab heavy chain, PaSta1 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUC
 
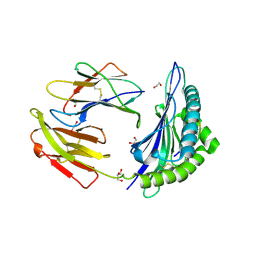 | | Crystal structure of HLA-B*44:05 (T73C) with 9mer EEFGRAFSF | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Jiang, J, Natarajan, K, Kim, E, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
1NZ6
 
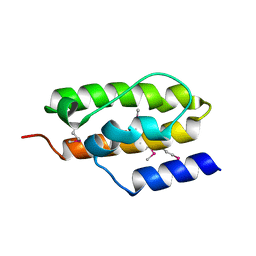 | | Crystal Structure of Auxilin J-Domain | | Descriptor: | Auxilin | | Authors: | Jiang, J, Taylor, A.B, Prasad, K, Ishikawa-Brush, Y, Hart, P.J, Lafer, E.M, Sousa, R. | | Deposit date: | 2003-02-16 | | Release date: | 2003-04-22 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface.
Biochemistry, 42, 2003
|
|
1P32
 
 | | CRYSTAL STRUCTURE OF HUMAN P32, A DOUGHNUT-SHAPED ACIDIC MITOCHONDRIAL MATRIX PROTEIN | | Descriptor: | MITOCHONDRIAL MATRIX PROTEIN, SF2P32 | | Authors: | Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1DVK
 
 | |
4X23
 
 | |
