4IFP
 
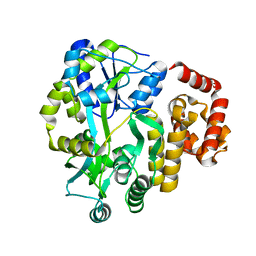 | | X-ray Crystal Structure of Human NLRP1 CARD Domain | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 1, ... | | Authors: | Jin, T, Curry, J, Smith, P, Jiang, J, Xiao, T. | | Deposit date: | 2012-12-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1.
Proteins, 81, 2013
|
|
4L19
 
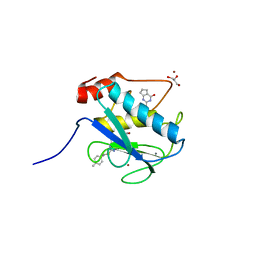 | | Matrix metalloproteinase-13 complexed with selective inhibitor compound Q1 | | Descriptor: | 2-[(4-methylbenzyl)sulfanyl]-3,5,6,7-tetrahydro-4H-cyclopenta[d]pyrimidin-4-one, CALCIUM ION, Collagenase 3, ... | | Authors: | Minond, D, Spicer, T.P, Jiang, J, Taylor, A.B, Hart, P.J, Roush, W.R, Fields, G.B, Hodder, P.S. | | Deposit date: | 2013-06-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Characterization of selective exosite-binding inhibitors of matrix metalloproteinase 13 that prevent articular cartilage degradation in vitro.
J.Med.Chem., 57, 2014
|
|
4LQC
 
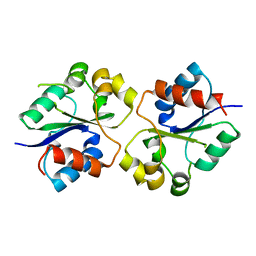 | | The crystal structures of the Brucella protein TcpB and the TLR adaptor protein TIRAP show structural differences in microbial TIR mimicry. | | Descriptor: | TcpB | | Authors: | Snyder, G.A, Smith, P, Fresquez, T, Cirl, C, Jiang, J, Snyder, N, Luchetti, T, Miethke, T, Xiao, T.S. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry.
J.Biol.Chem., 289, 2014
|
|
4LQD
 
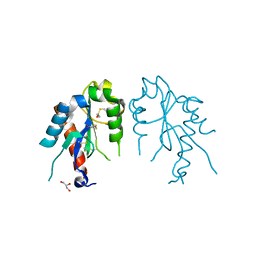 | | The crystal structures of the Brucella protein TcpB and the TLR adaptor protein TIRAP show structural differences in microbial TIR mimicry | | Descriptor: | GLYCEROL, Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Snyder, G.A, Smith, P, Jiang, J, Xiao, T.S. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry.
J.Biol.Chem., 289, 2014
|
|
5UN8
 
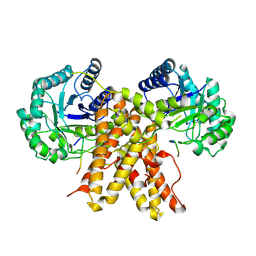 | |
5VKU
 
 | | An atomic structure of the human cytomegalovirus (HCMV) capsid with its securing layer of pp150 tegument protein | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | Authors: | Yu, X, Jih, J, Jiang, J, Zhou, H. | | Deposit date: | 2017-04-24 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150.
Science, 356, 2017
|
|
5TKE
 
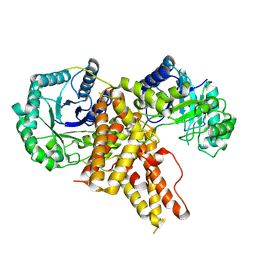 | | Crystal Structure of Eukaryotic Hydrolase | | Descriptor: | O-GlcNAcase: chimera construct | | Authors: | Li, B, Jiang, J. | | Deposit date: | 2016-10-06 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4IKM
 
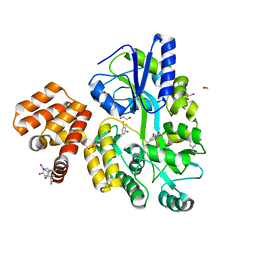 | | X-ray structure of CARD8 CARD domain | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Maltose-binding periplasmic protein, ... | | Authors: | Jin, T, Huang, M, Smith, P, Jiang, J, Xiao, T. | | Deposit date: | 2012-12-26 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4606 Å) | | Cite: | The structure of the CARD8 caspase-recruitment domain suggests its association with the FIIND domain and procaspases through adjacent surfaces.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4OI7
 
 | | RAGE recognizes nucleic acids and promotes inflammatory responses to DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*TP*AP*GP*CP*AP*TP*CP*GP*TP*TP*GP*CP*AP*G)-3', 5'-D(*CP*TP*GP*CP*AP*AP*CP*GP*AP*TP*GP*CP*TP*AP*CP*GP*AP*AP*CP*GP*TP*G)-3', ... | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
5VVX
 
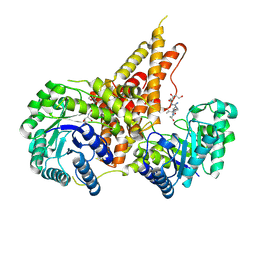 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lamin B1, Protein O-GlcNAcase | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
5G0M
 
 | | beta-glucuronidase with an activity-based probe (N-acyl cyclophellitol aziridine) bound | | Descriptor: | (1S,2R,3R,4S,6S)-6-[(8-azidooctanoyl)amino]-2,3,4-trihydroxycyclohexane-1-carboxylate, BETA-GLUCURONIDASE, PHOSPHATE ION | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-03-21 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of a Bacteria Beta-Glucuronidase in Gh79 Family
To be Published
|
|
5VVV
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, a-crystallin B | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
5UN9
 
 | | The crystal structure of human O-GlcNAcase in complex with Thiamet-G | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, Protein O-GlcNAcase | | Authors: | Li, B, Jiang, J. | | Deposit date: | 2017-01-30 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VVO
 
 | |
5VVU
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, TAB1 peptide | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
5VVT
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ELK1 peptide, Protein O-GlcNAcase | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
4OI8
 
 | | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. | | Descriptor: | 5'-D(*CP*CP*AP*TP*GP*AP*CP*TP*GP*TP*AP*GP*GP*AP*AP*AP*CP*TP*CP*TP*AP*GP*A)-3', 5'-D(*CP*TP*CP*TP*AP*GP*AP*GP*TP*TP*TP*CP*CP*TP*AP*CP*AP*GP*TP*CP*AP*TP*G)-3', Advanced glycosylation end product-specific receptor | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
7EBK
 
 | |
8G91
 
 | |
8G90
 
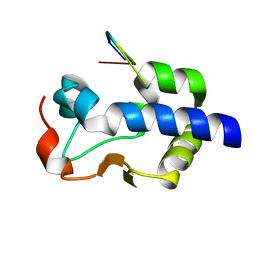 | |
5Z7D
 
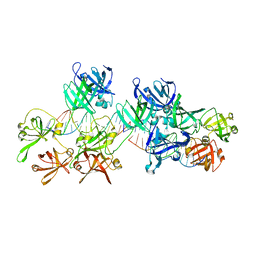 | | p204HINab-dsDNA complex structure | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*CP*AP*GP*AP*AP*AP*GP*AP*GP*AP*GP*C)-3'), Interferon-activable protein 204 | | Authors: | Jin, T, Jiang, J, Xiao, T.S. | | Deposit date: | 2018-01-28 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
7EBJ
 
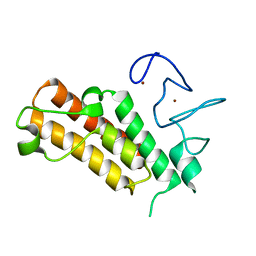 | | Apo structure of the mouse Trim66 PHD-Bromo dual domain | | Descriptor: | Tripartite motif-containing protein 66, ZINC ION | | Authors: | Wang, Z, Jiang, J. | | Deposit date: | 2021-03-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A TRIM66/DAX1/Dux axis suppresses the totipotent 2-cell-like state in murine embryonic stem cells.
Cell Stem Cell, 29, 2022
|
|
7CJG
 
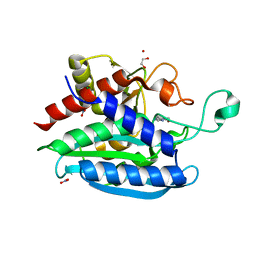 | | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | 5,6-DIMETHYLBENZIMIDAZOLE, GLYCEROL, Glutamine cyclotransferase-related protein, ... | | Authors: | Ruiz-Carrillo, D, Lamers, S, Feng, Q, Yu, S, Sun, B, Jiang, J, Lukman, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase.
Biol.Chem., 402, 2021
|
|
7C72
 
 | | Structure of a mycobacterium tuberculosis puromycin-hydrolyzing peptidase | | Descriptor: | D-MALATE, GLYCEROL, Prolyl oligopeptidase | | Authors: | Ruiz-Carrillo, D, Zhao, Y.H, Feng, Q, Zhou, X, Zhang, Y, Jiang, J, Lukman, M. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.00004458 Å) | | Cite: | Mycobacterium tuberculosis puromycin hydrolase displays a prolyl oligopeptidase fold and an acyl aminopeptidase activity.
Proteins, 89, 2021
|
|
7CJE
 
 | | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | GLYCEROL, Glutamine cyclotransferase-related protein, MAGNESIUM ION, ... | | Authors: | Ruiz-Carrillo, D, Lamers, S, Feng, Q, Yu, S, Sun, B, Jiang, J, Lukman, M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.950007 Å) | | Cite: | Structural and kinetic characterization of Porphyromonas gingivalis glutaminyl cyclase.
Biol.Chem., 402, 2021
|
|
