4PZV
 
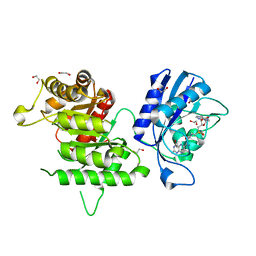 | | Crystal structure of Francisella tularensis HPPK-DHPS in complex with bisubstrate analog HPPK inhibitor J1D | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, 5'-{[2-({N-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]glycyl}amino)ethyl]sulfonyl}-5'-deoxyadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structural enzymology and inhibition of the bi-functional folate pathway enzyme HPPK-DHPS from the biowarfare agent Francisella tularensis.
Febs J., 281, 2014
|
|
6AN6
 
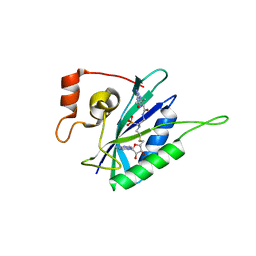 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate analogue inhibitor HP-72 | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-{2-[{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}(phosphonomethyl)amino]ethyl}-5'-thioadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bisubstrate analogue inhibitors of HPPK: Transition state mimetics
to be published
|
|
6AN4
 
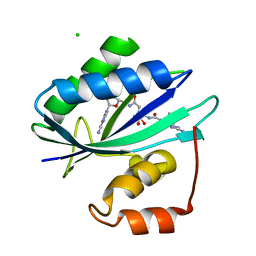 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate analogue inhibitor HP-39 (J1F) | | Descriptor: | ((2-(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carboxamido)-N-(2-((((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)amino)-2-oxoethyl)acetamido)methyl)phosphonic acid, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bisubstrate analog inhibitors of HPPK: Transition state mimetics
to be published
|
|
2HHD
 
 | | OXYGEN AFFINITY MODULATION BY THE N-TERMINI OF THE BETA-CHAINS IN HUMAN AND BOVINE HEMOGLOBIN | | Descriptor: | HEMOGLOBIN (DEOXY) (ALPHA CHAIN), HEMOGLOBIN (DEOXY) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gilliland, G.L, Pechik, I, Fronticelli, C, Ji, X. | | Deposit date: | 1994-09-29 | | Release date: | 1995-01-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic, molecular modeling, and biophysical characterization of the valine beta 67 (E11)-->threonine variant of hemoglobin.
Biochemistry, 35, 1996
|
|
2HK8
 
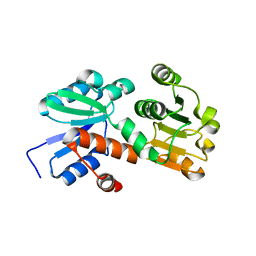 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus at 2.35 angstrom resolution | | Descriptor: | Shikimate dehydrogenase | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
2HK9
 
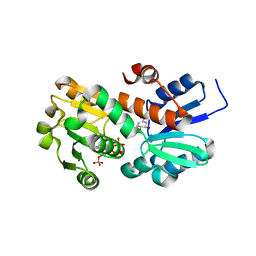 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with shikimate and NADP+ at 2.2 angstrom resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
2HK7
 
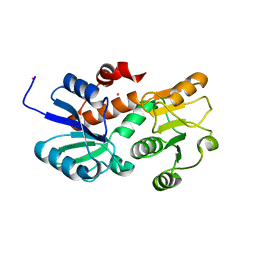 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with mercury at 2.5 angstrom resolution | | Descriptor: | MERCURY (II) ION, Shikimate dehydrogenase | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
6AP9
 
 | |
1AJ9
 
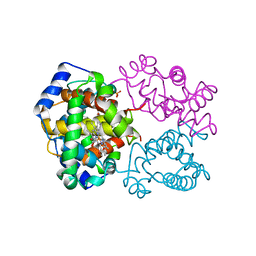 | | R-STATE HUMAN CARBONMONOXYHEMOGLOBIN ALPHA-A53S | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), ... | | Authors: | Vasquez, G.B, Ji, X, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1997-05-16 | | Release date: | 1998-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human carboxyhemoglobin at 2.2 A resolution: structure and solvent comparisons of R-state, R2-state and T-state hemoglobins.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1NPR
 
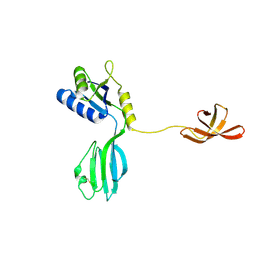 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN C222(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzhan, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
1NPP
 
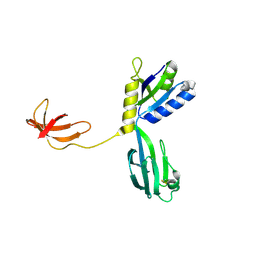 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN P2(1) | | Descriptor: | ISOPROPYL ALCOHOL, Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzahn, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
3FTD
 
 | |
3FTF
 
 | | Crystal structure of A. aeolicus KsgA in complex with RNA and SAH | | Descriptor: | 5'-R(P*AP*AP*CP*CP*GP*UP*AP*GP*GP*GP*GP*AP*AP*CP*CP*UP*GP*CP*GP*GP*UP*U)-3', Dimethyladenosine transferase, POTASSIUM ION, ... | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-01-12 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Binding of RNA and Cofactor by a KsgA Methyltransferase.
Structure, 17, 2009
|
|
3DJ9
 
 | | Crystal Structure of an isolated, unglycosylated antibody CH2 domain | | Descriptor: | Ig gamma-1 chain C region | | Authors: | Prabakaran, P, Vu, B.K, Gan, J, Dimitrov, D.S, Ji, X. | | Deposit date: | 2008-06-22 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an isolated unglycosylated antibody C(H)2 domain.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3FTC
 
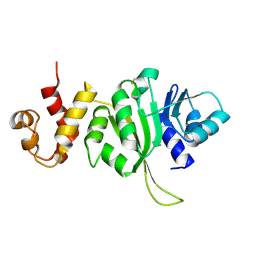 | |
1B48
 
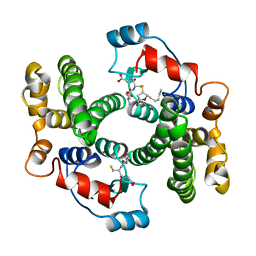 | |
3FTE
 
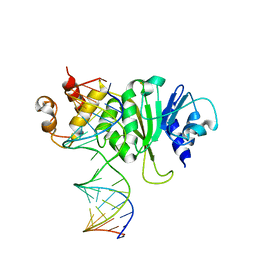 | | Crystal structure of A. aeolicus KsgA in complex with RNA | | Descriptor: | 5'-R(P*AP*AP*CP*CP*GP*UP*AP*GP*GP*GP*GP*AP*AP*CP*CP*UP*GP*CP*GP*GP*UP*U)-3', Dimethyladenosine transferase | | Authors: | Tu, C, Ji, X. | | Deposit date: | 2009-01-12 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Binding of RNA and Cofactor by a KsgA Methyltransferase.
Structure, 17, 2009
|
|
1ESR
 
 | |
1RHH
 
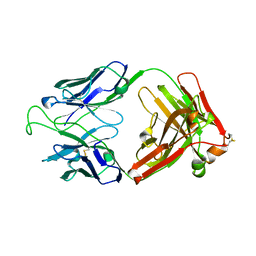 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
1EX6
 
 | |
1EX7
 
 | |
3ILL
 
 | | Crystal structure of E. coli HPPK(D97A) | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional roles of residues D95 and D97 in E. coli HPPK
To be Published
|
|
3ILI
 
 | | Crystal structure of E. coli HPPK(D95A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional roles of residues D95 and D97 in E. coli HPPK
To be Published
|
|
1I4S
 
 | |
3HSZ
 
 | | Crystal structure of E. coli HPPK(F123A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Pterin-binding site mutation Y53A, N55A or F123A and activity of E. coli HPPK
To be Published
|
|
