6X4I
 
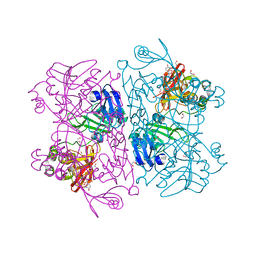 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate | | Descriptor: | 1,2-ETHANEDIOL, 3'-URIDINEMONOPHOSPHATE, SODIUM ION, ... | | Authors: | Chang, C, Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WY4
 
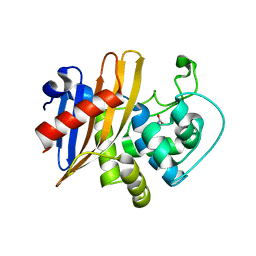 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-12 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
6WEN
 
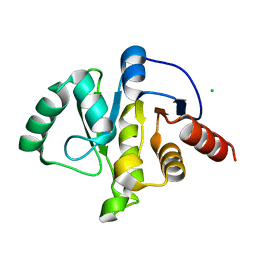 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form | | Descriptor: | CHLORIDE ION, Non-structural protein 3 | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-02 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-11 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
4ZXW
 
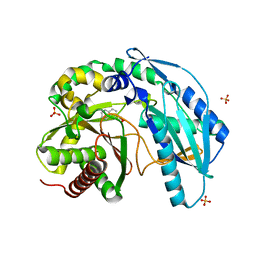 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | Descriptor: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
7SUE
 
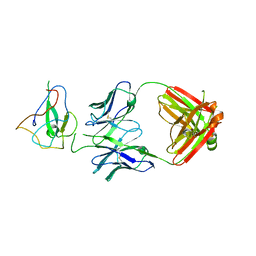 | | Crystal Structure of Human Fab S24-188 in the complex with the N-teminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | Nucleoprotein, S24-188 Fab Heavy chain, S24-188 Fab Light chain | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7STS
 
 | | Crystal Structure of Human Fab S24-1379 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | Fab S24-1379, heavy chain, light chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7STR
 
 | | Crystal Structure of Human Fab S24-1063 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Fab S24-1063, Heavy chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
5IR2
 
 | | Crystal structure of novel cellulases from microbes associated with the gut ecosystem | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cellulase, ... | | Authors: | Chang, C, Mack, J, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-11 | | Release date: | 2016-03-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Crystal structure of novel cellulases from microbes associated with the gut ecosystem
To Be Published
|
|
6MGY
 
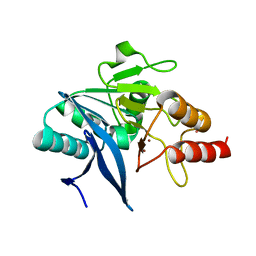 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae | | Descriptor: | GLYCEROL, Metallo-beta-lactamase NDM-5, POTASSIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae
To Be Published
|
|
6MH0
 
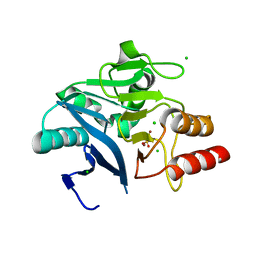 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 3 from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Metallo-beta-lactamase NDM-3, SULFATE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 3 from Klebsiella pneumoniae
To Be Published
|
|
6NJC
 
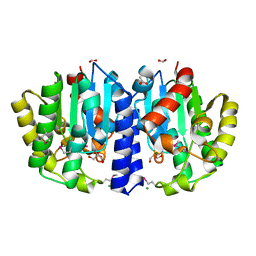 | | Crystal Structure of the Sialate O-acetylesterase from Bacteroides vulgatus | | Descriptor: | ACETIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Li, H, Biglow, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-01-03 | | Release date: | 2019-01-16 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Sialate O-acetylesterase from Bacteroides vulgatus
To Be Published
|
|
6NKF
 
 | | Crystal Structure of the Lipase Lip_vut4 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.232 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut4 from Goat Rumen metagenome.
To Be Published
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5EV7
 
 | | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames | | Descriptor: | Conserved domain protein | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The crystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames.
To Be Published
|
|
5F1Y
 
 | |
5FCD
 
 | | Crystal structure of MccD protein | | Descriptor: | CHLORIDE ION, MccD, UNK-UNK-UNK-MSE-UNK, ... | | Authors: | Nocek, B, Jedrzejczak, R, Anderson, W.F, Severinov, K, Dubiley, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MccD protein.
To Be Published
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
5JBR
 
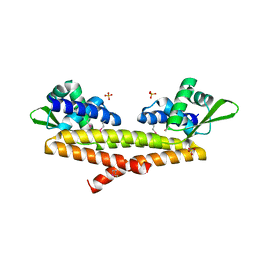 | | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae | | Descriptor: | SULFATE ION, Uncharacterized protein Bcav_2135 | | Authors: | Chang, C, Cuff, M, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-04-13 | | Release date: | 2016-04-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae
To Be Published
|
|
5JMB
 
 | |
3D0K
 
 | | Crystal structure of the LpqC, poly(3-hydroxybutyrate) depolymerase from Bordetella parapertussis | | Descriptor: | CHLORIDE ION, FORMIC ACID, Putative poly(3-hydroxybutyrate) depolymerase LpqC, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the LpqC, Poly(3-hydroxybutyrate) Depolymerase from Bordetella parapertussis.
To be Published
|
|
3D3S
 
 | | Crystal structure of L-2,4-diaminobutyric acid acetyltransferase from Bordetella parapertussis | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, GLYCEROL, L-2,4-diaminobutyric acid acetyltransferase, ... | | Authors: | Kim, Y, Volkart, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of L-2,4-diaminobutyric acid acetyltransferase from Bordetella parapertussis.
To be Published
|
|
3DO8
 
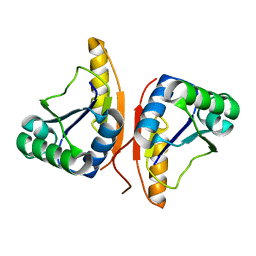 | |
