6WXC
 
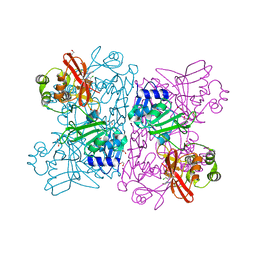 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
5VVI
 
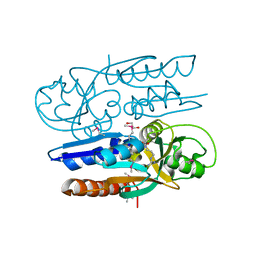 | | Crystal Structure of the Ligand Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens in the Complex with Octopine | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
3TY7
 
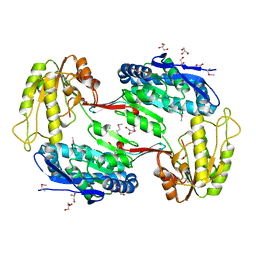 | | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Joachimiak, G, Jedrzejczak, R, Rubin, E, Ioerger, T, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus
To be Published
|
|
4IYI
 
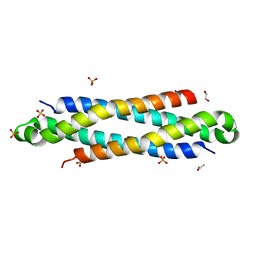 | | The crystal structure of a secreted protein EsxB (wild-type, C-term. His-tagged) from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | The crystal structure of a secreted protein EsxB (wild-type, C-term. His-tagged) from Bacillus anthracis str. Sterne
To be Published
|
|
4IYH
 
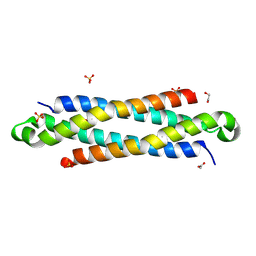 | | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne
To be Published
|
|
4IR0
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6W6Y
 
 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, Non-structural protein 3 | | Authors: | Michalska, K, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3RKJ
 
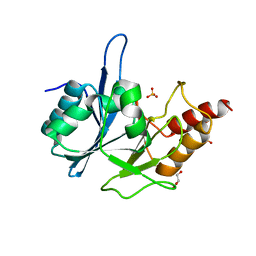 | | Crystal Structure of New Delhi Metallo-Beta-Lactamase-1 from Klebsiella pnueumoniae | | Descriptor: | Beta-lactamase NDM-1, GLYCEROL, SULFATE ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Binkowski, T.A, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3RKK
 
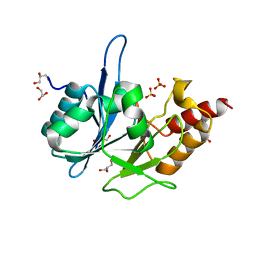 | | Crystal Structure of New Delhi Metallo-Beta-Lactamase-1 from Klebsiella pneumoniae | | Descriptor: | ACETIC ACID, Beta-lactamase NDM-1, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Binkowski, T.A, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3RRL
 
 | | Complex structure of 3-oxoadipate coA-transferase subunit A and B from Helicobacter pylori 26695 | | Descriptor: | GLYCEROL, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit A, Succinyl-CoA:3-ketoacid-coenzyme A transferase subunit B | | Authors: | Nocek, B, Stein, A, Marshall, N, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Complex structure of 3-oxoadipate coA-transferase subunit A and B
from Helicobacter pylori 26695
TO BE PUBLISHED
|
|
3RPF
 
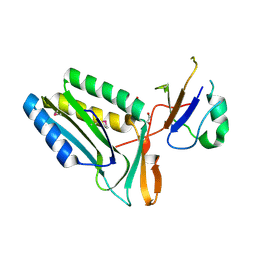 | | Protein-protein complex of subunit 1 and 2 of Molybdopterin-converting factor from Helicobacter pylori 26695 | | Descriptor: | 1,2-ETHANEDIOL, Molybdopterin converting factor, subunit 1 (MoaD), ... | | Authors: | Nocek, B, Stein, A, Marshall, N, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-26 | | Release date: | 2011-06-29 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein-protein complex of subunit 1 and 2 of Molybdopterin-converting factor from Helicobacter pylori 26695
TO BE PUBLISHED
|
|
3SBL
 
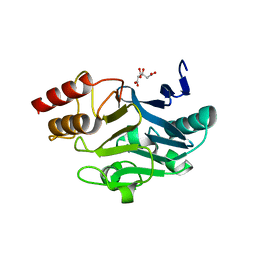 | | Crystal Structure of New Delhi Metal-beta-lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CITRIC ACID | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
6UE2
 
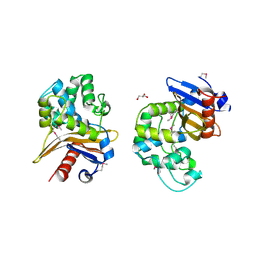 | | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630.
To Be Published
|
|
4J11
 
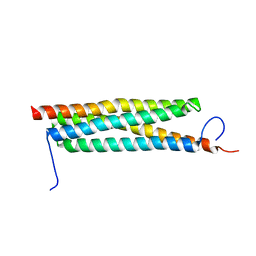 | | The crystal structure of a secreted protein ESXB (wild-type, in P21 space group) from Bacillus anthracis str. sterne | | Descriptor: | SECRETED PROTEIN ESXB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Crystal Structure of a Secreted Protein Esxb (Wild-Type, in P21 Space Group) from Bacillus Anthracis Str. Sterne
To be Published
|
|
4J10
 
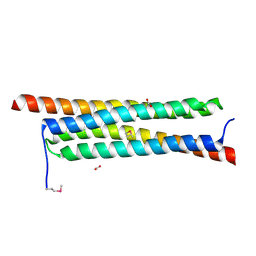 | | The crystal structure of a secreted protein ESXB (SeMet-labeled) from Bacillus anthracis str. Sterne | | Descriptor: | FORMIC ACID, SECRETED PROTEIN ESXB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Crystal Structure of a Secreted Protein Esxb (Semet-Labeled) from Bacillus Anthracis Str. Sterne
To be Published
|
|
4J41
 
 | | The crystal structure of a secreted protein EsxB (Mutant P67A) from Bacillus anthracis str. Sterne | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-06 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.522 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant P67A) from Bacillus anthracis str. Sterne
To be Published
|
|
7KAB
 
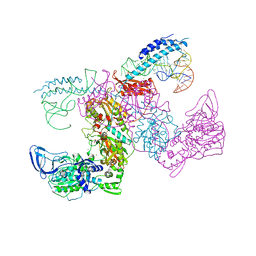 | | M. tuberculosis PheRS complex with cognate precursor tRNA and phenylalanine | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHENYLALANINE, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Wower, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
4J42
 
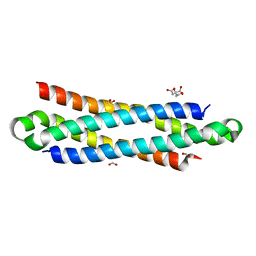 | | The crystal structure of a secreted protein EsxB (Mutant Y65F) from Bacillus anthracis str. Sterne | | Descriptor: | CITRATE ANION, FORMIC ACID, secreted protein EsxB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-06 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant Y65F) from Bacillus anthracis str. Sterne
To be Published
|
|
4J7J
 
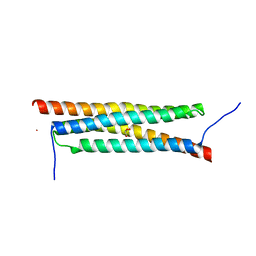 | | The crystal structure of a secreted protein EsxB (Mutant G53A) from Bacillus anthracis str. Sterne | | Descriptor: | GLYCEROL, secreted protein EsxB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.458 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant, G53A) from Bacillus anthracis str. Sterne
To be Published
|
|
4J7K
 
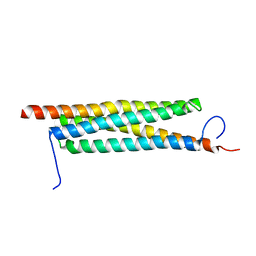 | | The crystal structure of a secreted protein EsxB (Mutant E54Q) from Bacillus anthracis str. Sterne | | Descriptor: | secreted protein EsxB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant E54Q) from Bacillus anthracis str. Sterne
To be Published
|
|
6W9C
 
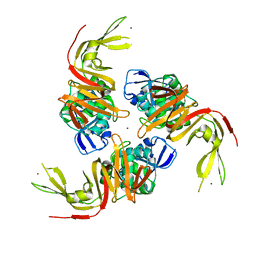 | | The crystal structure of papain-like protease of SARS CoV-2 | | Descriptor: | CHLORIDE ION, Non-structural protein 3, ZINC ION | | Authors: | Osipiuk, J, Jedrzejczak, R, Tesar, C, Endres, M, Stols, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-22 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of papain-like protease of SARS CoV-2
to be published
|
|
6WY4
 
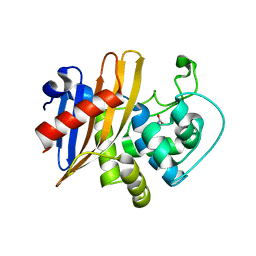 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-12 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
