7STR
 
 | | Crystal Structure of Human Fab S24-1063 in the Complex with the N-teminal Domain of Nucleocapsid Protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Fab S24-1063, Heavy chain, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-11-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
4Q52
 
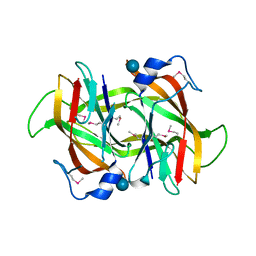 | | 2.60 Angstrom resolution crystal structure of a conserved uncharacterized protein from Chitinophaga pinensis DSM 2588 | | Descriptor: | Uncharacterized protein, beta-D-glucopyranose | | Authors: | Halavaty, A.S, Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Minasov, G, Jedrzejczak, R, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom resolution crystal structure of a conserved uncharacterized protein from Chitinophaga pinensis DSM 2588
To be Published
|
|
5L07
 
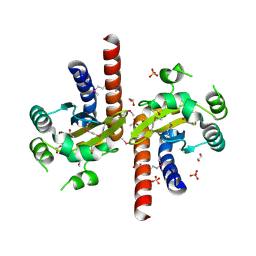 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Quorum-sensing transcriptional activator, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
4QVS
 
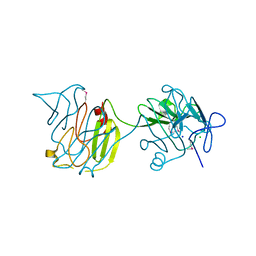 | | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405 | | Descriptor: | CHLORIDE ION, S-layer domain-containing protein, SODIUM ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405
To be Published
|
|
4QYB
 
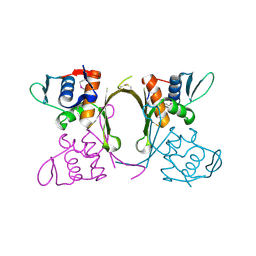 | | 2.1 Angstrom resolution crystal structure of uncharacterized protein, disulfide-bridged dimer, from Burkholderia cenocepacia J2315 | | Descriptor: | Uncharacterized protein | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of uncharacterized protein, disulfide-bridged dimer, from Burkholderia cenocepacia J2315
To be Published
|
|
6DIN
 
 | | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase | | Descriptor: | SULFATE ION, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Chang, C, Jedrzejczak, R, Chhor, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-23 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase
To Be Published
|
|
5L09
 
 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica in complex with 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide, ACETIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5L10
 
 | | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winan, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-07-28 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia.
To Be Published
|
|
3DNP
 
 | | Crystal structure of Stress response protein yhaX from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Stress response protein yhaX | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Stress response protein yhaX from Bacillus subtilis
To be Published
|
|
6MGZ
 
 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NDM-4, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae
To Be Published
|
|
6MGX
 
 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae | | Descriptor: | Metallo-beta-lactamase, SULFATE ION, ZINC ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 6 Klebsiella pneumoniae
To Be Published
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | Authors: | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-07 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
6C9Z
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | Descriptor: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, Glycosyl hydrolase, family 31 | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) W169Y mutant FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
6CA3
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
6C9X
 
 | | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose | | Descriptor: | (1S,2S,3R,4S,5S)-5-[(1,3-dihydroxypropan-2-yl)amino]-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.457 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE alpha-Glucosidase (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with voglibose
To Be Published
|
|
6CA1
 
 | | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, GLYCEROL, Glycosyl hydrolase, ... | | Authors: | Tan, K, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Macromolecular Research (MCMR) | | Deposit date: | 2018-01-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | THE CRYSTAL STRUCTURE OF THE W169Y MUTANT OF ALPHA-GLUCOSIDASE (GH 31) FROM RUMINOCOCCUS OBEUM ATCC 29174 in complex with miglitol
To Be Published
|
|
5KCK
 
 | | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4 | | Descriptor: | Anthranilate synthase component I, GLYCEROL | | Authors: | Chang, C, Michalska, K, Bigelow, L, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-06 | | Release date: | 2016-06-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4
To Be Published
|
|
7KB3
 
 | | The structure of a sensor domain of a histidine kinase (VxrA) from Vibrio cholerae O1 biovar eltor str. N16961, 2nd form | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Wu, R, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 2021
|
|
6NKC
 
 | | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome.
To Be Published
|
|
6NKG
 
 | | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Lip_vut5, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome.
To Be Published
|
|
6NKD
 
 | | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome.
To Be Published
|
|
4Q31
 
 | | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-07 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural dynamics of a methionine gamma-lyase for calicheamicin biosynthesis: Rotation of the conserved tyrosine stacking with pyridoxal phosphate.
Struct Dyn, 3, 2016
|
|
3GKU
 
 | | Crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940 | | Descriptor: | Probable RNA-binding protein | | Authors: | Tan, K, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a probable RNA-binding protein from Clostridium symbiosum ATCC 14940
To be Published
|
|
3GYG
 
 | | Crystal structure of yhjK (haloacid dehalogenase-like hydrolase protein) from Bacillus subtilis | | Descriptor: | MAGNESIUM ION, NTD biosynthesis operon putative hydrolase ntdB | | Authors: | Nocek, B, Stein, A, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of yhjK (haloacid dehalogenase-like hydrolase protein) from Bacillus subtilis
To be Published
|
|
3H1Q
 
 | | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ethanolamine utilization protein EutJ | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Kinney, J, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-13 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutJ from Carboxydothermus hydrogenoformans
To be Published
|
|
