8PA2
 
 | |
8PA1
 
 | |
1MPX
 
 | | ALPHA-AMINO ACID ESTER HYDROLASE LABELED WITH SELENOMETHIONINE | | Descriptor: | CALCIUM ION, GLYCEROL, alpha-amino acid ester hydrolase | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Hensgens, C.M.H, de Vries, E.J, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2002-09-13 | | Release date: | 2003-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The sequence and crystal structure of the alpha-amino acid ester hydrolase from Xanthomonas citri define a new family of beta-lactam antibiotic acylases.
J.Biol.Chem., 278, 2003
|
|
1EHY
 
 | | X-ray structure of the epoxide hydrolase from agrobacterium radiobacter ad1 | | Descriptor: | POTASSIUM ION, PROTEIN (SOLUBLE EPOXIDE HYDROLASE) | | Authors: | Nardini, M, Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Rink, R, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 1998-10-17 | | Release date: | 1999-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The x-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides.
J.Biol.Chem., 274, 1999
|
|
8BIT
 
 | | Crystal structure of acyl-CoA synthetase from Metallosphaera sedula in complex with Coenzyme A and acetyl-AMP | | Descriptor: | 4-hydroxybutyrate--CoA ligase 1, COENZYME A, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2022-11-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
8BIQ
 
 | | Crystal structure of acyl-COA synthetase from Metallosphaera sedula in complex with acetyl-AMP | | Descriptor: | 4-hydroxybutyrate--CoA ligase 1, ADENOSINE MONOPHOSPHATE, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2022-11-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
6YRA
 
 | |
6Z1X
 
 | |
6Z1W
 
 | |
5AC3
 
 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
2B4K
 
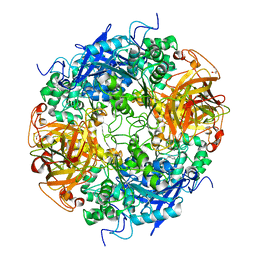 | | Acetobacter turbidans alpha-amino acid ester hydrolase complexed with phenylglycine | | Descriptor: | Alpha-amino acid ester hydrolase, D-PHENYLGLYCINE, GLYCEROL | | Authors: | Barends, T.R.M, Polderman-Tijmes, J.J, Jekel, P.A, Williams, C, Wybenga, G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-09-26 | | Release date: | 2005-12-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Acetobacter turbidans alpha-amino acid ester hydrolase: how a single mutation improves an antibiotic-producing enzyme.
J.Biol.Chem., 281, 2006
|
|
8RJ1
 
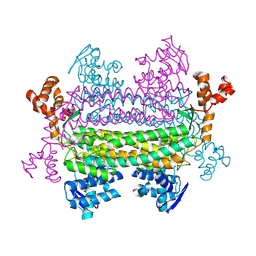 | |
8RJ0
 
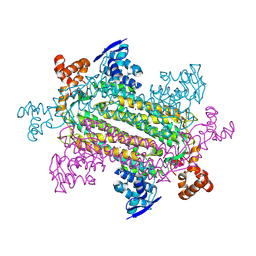 | |
4C5U
 
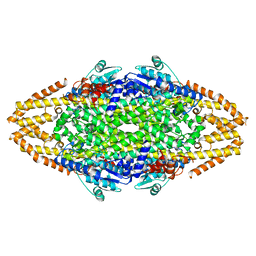 | | Structural Investigations into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus chinensis | | Descriptor: | PHENYLALANINE AMMONIA-LYASE | | Authors: | Wybenga, G.G, Szymanski, W, Wu, B, Feringa, B.L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Investigations Into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus Chinensis.
Biochemistry, 53, 2014
|
|
7B4I
 
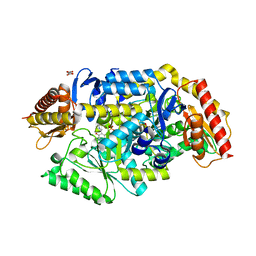 | | Thermostable omega transaminase PjTA-R6 variant W58G engineered for asymmetric synthesis of enantiopure bulky amines | | Descriptor: | Aspartate aminotransferase family protein, PYRIDOXAL-5'-PHOSPHATE, SUCCINIC ACID | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2020-12-02 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
7B4J
 
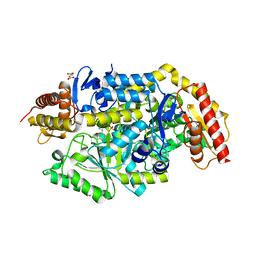 | | Thermostable omega transaminase PjTA-R6 variant W58M/F86L/R417L engineered for asymmetric synthesis of enantiopure bulky amines | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase family protein, SUCCINIC ACID | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2020-12-02 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
4C5R
 
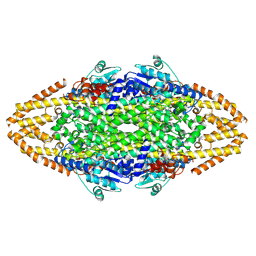 | | Structural Investigations into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus chinensis | | Descriptor: | (3S)-3-amino-2,2-difluoro-3-phenylpropanoic acid, GLYCEROL, PHENYLALANINE AMMONIA-LYASE | | Authors: | Wybenga, G.G, Szymanski, W, Wu, B, Feringa, B.L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Investigations Into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus Chinensis.
Biochemistry, 53, 2014
|
|
1FXH
 
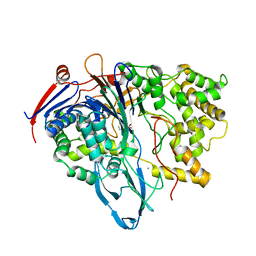 | | MUTANT OF PENICILLIN ACYLASE IMPAIRED IN CATALYSIS WITH PHENYLACETIC ACID IN THE ACTIVE SITE | | Descriptor: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN ACYLASE | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-26 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
1FXV
 
 | | PENICILLIN ACYLASE MUTANT IMPAIRED IN CATALYSIS WITH PENICILLIN G IN THE ACTIVE SITE | | Descriptor: | CALCIUM ION, PENICILLIN ACYLASE, PENICILLIN G | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-27 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
4C5S
 
 | | Structural Investigations into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus chinensis | | Descriptor: | (3S)-3-amino-2,2-difluoro-3-phenylpropanoic acid, PHENYLALANINE AMMONIA-LYASE | | Authors: | Wybenga, G.G, Szymanski, W, Wu, B, Feringa, B.L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Investigations Into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus Chinensis.
Biochemistry, 53, 2014
|
|
4CQ5
 
 | | Structural Investigations into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus chinensis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHENYLALANINE AMINOMUTASE, ... | | Authors: | Wybenga, G.G, Szymanski, W, Wu, B, Feringa, B.L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2014-02-11 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Investigations Into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus Chinensis.
Biochemistry, 53, 2014
|
|
4C6G
 
 | | Structural Investigations into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus chinensis | | Descriptor: | PHENYLALANINE AMMONIA-LYASE | | Authors: | Wybenga, G.G, Szymanski, W, Wu, B, Feringa, B.L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigations Into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus Chinensis.
Biochemistry, 53, 2014
|
|
4R9K
 
 | | Structure of thermostable eightfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis | | Descriptor: | (2R)-2-hydroxyhexanamide, GLYCEROL, Limonene-1,2-epoxide hydrolase | | Authors: | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
4R9L
 
 | | Structure of a thermostable elevenfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis, containing two stabilizing disulfide bonds | | Descriptor: | (2R)-2-hydroxyhexanamide, Limonene-1,2-epoxide hydrolase | | Authors: | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
4IXT
 
 | | Structure of a 37-fold mutant of halohydrin dehalogenase (HheC) bound to ethyl (R)-4-cyano-3-hydroxybutyrate | | Descriptor: | CHLORIDE ION, Halohydrin dehalogenase, ethyl (3R)-4-cyano-3-hydroxybutanoate | | Authors: | Floor, R.J, Schallmey, M, Hauer, B, Breuer, M, Jekel, P.A, Wijma, H.J, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Biocatalytic and structural properties of a highly engineered halohydrin dehalogenase.
Chembiochem, 14, 2013
|
|
