8FIK
 
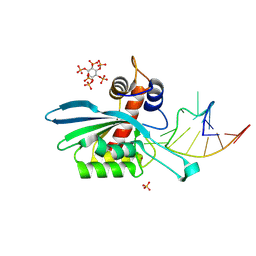 | | APOBEC3A E72A inactive mutant in complex with ATTC-hairpin DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*CP*CP*CP*AP*TP*CP*AP*TP*TP*CP*GP*AP*TP*GP*GP*G)-3'), DNA dC->dU-editing enzyme APOBEC-3A, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
8FII
 
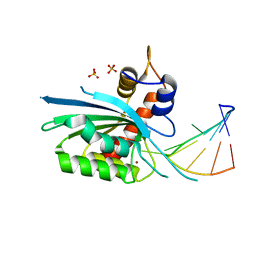 | | Wild type APOBEC3A in complex with TT(FdZ)-hairpin inhibitor (crystal form 1) | | Descriptor: | DNA (5'-D(P*GP*CP*GP*CP*TP*TP*(UFP)P*GP*CP*GP*C)-3'), DNA dC->dU-editing enzyme APOBEC-3A, PHOSPHATE ION, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
8FIJ
 
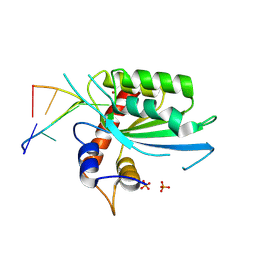 | | Wild type APOBEC3A in complex with TT(FdZ)-hairpin inhibitor (crystal form 2) | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*TP*TP*(UFP)P*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, PHOSPHATE ION, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
8FIM
 
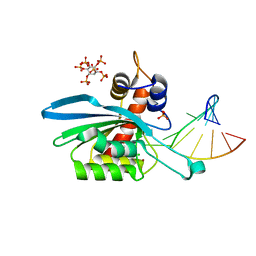 | | Structure of APOBEC3A (E72A inactive mutant) in complex with TTC-hairpin DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*TP*GP*CP*GP*CP*TP*TP*CP*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
2YPO
 
 | | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with phenylalanine bound in only one site | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Blackmore, N.J, Reichau, S, Jiao, W, Hutton, R.D, Baker, E.N, Jameson, G.B, Parker, E.J. | | Deposit date: | 2012-10-31 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three Sites and You are Out: Ternary Synergistic Allostery Controls Aromatic Aminoacid Biosynthesis in Mycobacterium Tuberculosis.
J.Mol.Biol., 425, 2013
|
|
3BLG
 
 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 6.2 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, H.M, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
3DU0
 
 | | E. coli dihydrodipicolinate synthase with first substrate, pyruvate, bound in active site | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Dobson, R.C.J, Devenish, S.R.A, Gerrard, J.A, Jameson, G.B. | | Deposit date: | 2008-07-16 | | Release date: | 2008-11-18 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The high-resolution structure of dihydrodipicolinate synthase from Escherichia coli bound to its first substrate, pyruvate.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
5UXN
 
 | | Type II DAH7PS from Pseudomonas aeruginosa with Tyr bound | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Sterritt, O.W, Jameson, G.B, Parker, E.J. | | Deposit date: | 2017-02-23 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Pseudoisostructural Type II DAH7PS Enzyme from Pseudomonas aeruginosa: Alternative Evolutionary Strategies to Control Shikimate Pathway Flux.
Biochemistry, 57, 2018
|
|
5UXM
 
 | | Type II DAH7PS from Pseudomonas aeruginosa with Trp bound | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Sterritt, O.W, Jameson, G.B, Parker, E.J. | | Deposit date: | 2017-02-23 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A Pseudoisostructural Type II DAH7PS Enzyme from Pseudomonas aeruginosa: Alternative Evolutionary Strategies to Control Shikimate Pathway Flux.
Biochemistry, 57, 2018
|
|
5UXO
 
 | | Type II DAH7PS from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Sterritt, O.W, Jameson, G.B, Parker, E.J. | | Deposit date: | 2017-02-23 | | Release date: | 2018-02-28 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Pseudoisostructural Type II DAH7PS Enzyme from Pseudomonas aeruginosa: Alternative Evolutionary Strategies to Control Shikimate Pathway Flux.
Biochemistry, 57, 2018
|
|
6BMC
 
 | | The structure of a dimeric type II DAH7PS associated with pyocyanin biosynthesis in Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Sterritt, O.W, Jameson, G.B, Parker, E.J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterisation of the entry point to pyocyanin biosynthesis inPseudomonas aeruginosadefines a new 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase subclass.
Biosci. Rep., 38, 2018
|
|
6BQI
 
 | | Structure of two-domain translational regulator Yih1 reveals a possible mechanism of action | | Descriptor: | Protein IMPACT homolog | | Authors: | Harjes, E, Jameson, G.B, Edwards, P.J.B, Goroncy, A.K, Loo, T, Norris, G.E. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Experimentally based structural model of Yih1 provides insight into its function in controlling the key translational regulator Gcn2.
Febs Lett., 2020
|
|
2AKQ
 
 | | The structure of bovine B-lactoglobulin A in crystals grown at very low ionic strength | | Descriptor: | Beta-lactoglobulin variant A | | Authors: | Adams, J.J, Anderson, B.F, Norris, G.E, Creamer, L.K, Jameson, G.B. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bovine beta-lactoglobulin (variant A) at very low ionic strength
J.Struct.Biol., 154, 2006
|
|
3I7Q
 
 | | Dihydrodipicolinate synthase mutant - K161A | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
3I7R
 
 | | Dihydrodipicolinate synthase - K161R | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
3I7S
 
 | | Dihydrodipicolinate synthase mutant - K161A - with the substrate pyruvate bound in the active site. | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
1EN6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Q146L MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1EN5
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Y34F MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
1EN4
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE E. COLI MANGANESE SUPEROXIDE DISMUTASE Q146H MUTANT | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Whittaker, M.M, Baker, E.N, Whittaker, J.W, Jameson, G.B. | | Deposit date: | 2000-03-20 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Outer sphere mutations perturb metal reactivity in manganese superoxide dismutase.
Biochemistry, 40, 2001
|
|
3NUD
 
 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from mycobacterium tuberculosis complexed with phenylalanine | | Descriptor: | PHENYLALANINE, PHOSPHATE ION, Probable 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Webby, C.J, Baker, E.N, Baker, H.M. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
3NV8
 
 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase in complex with phosphoenol pyruvate and manganese (thesit-free) | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Hutton, R.H, Webby, C.J, Baker, E.N, Baker, H.M. | | Deposit date: | 2010-07-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
3NUE
 
 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from mycobacterium tuberculosis complexed with tryptophan | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Webby, C.J, Baker, E.N, Baker, H.M. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
3PB0
 
 | |
3PB2
 
 | |
1MA1
 
 | | Structure and properties of the atypical iron superoxide dismutase from Methanobacterium thermoautotrophicum | | Descriptor: | FE (III) ION, superoxide dismutase | | Authors: | Adams, J.J, Anderson, B.F, Renault, J.P, Verchere-Beaur, C, Morgenstern-Badarau, I, Jameson, G.B. | | Deposit date: | 2002-07-31 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and properties of the atypical iron superoxide dismutase from Methanobacterium thermoautotrophicum
To be published
|
|
