2X0A
 
 | |
2V0B
 
 | |
4FON
 
 | |
2V8B
 
 | |
4RBR
 
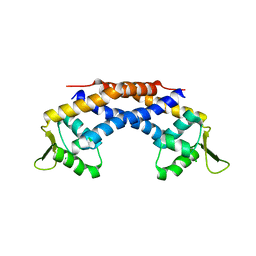 | | Crystal structure of Repressor of Toxin (Rot), a central regulator of Staphylococcus aureus virulence | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator rot | | Authors: | Killikelly, A, Jakoncic, J, Sampson, J.M, Kong, X.-P. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Functional Characterization of Repressor of Toxin (Rot), a Central Regulator of Staphylococcus aureus Virulence.
J.Bacteriol., 197, 2015
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MRR
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Leupeptin | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, LEUPEPTIN | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MGU
 
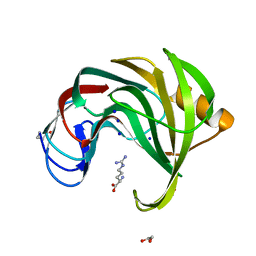 | |
5D14
 
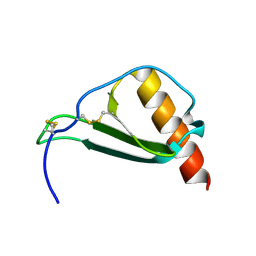 | |
1UWM
 
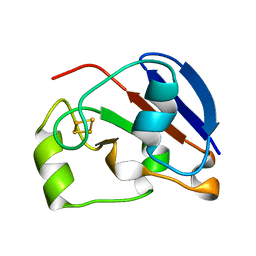 | | reduced ferredoxin 6 from Rhodobacter capsulatus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN VI | | Authors: | Sainz, G, Jakoncic, J, Sieker, L.C, Stojanoff, V, Sanishvili, N, Asso, M, Bertrand, P, Armengaud, J, Jouanneau, Y. | | Deposit date: | 2004-02-05 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a [2Fe-2S] Ferredoxin from Rhodobacter Capsulatus Likely Involved in Fe-S Cluster Biogenesis and Conformational Changes Observed Upon Reduction.
J.Biol.Inorg.Chem., 11, 2006
|
|
4OVN
 
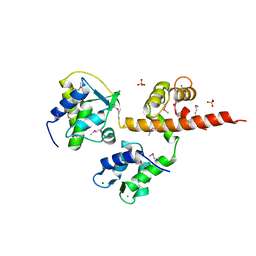 | | Voltage-gated Sodium Channel 1.5 (Nav1.5) C-terminal domain in complex with Calmodulin poised for activation | | Descriptor: | Calmodulin, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Boto, A, Jakoncic, J, Tomaselli, G.F, Amzel, L.M. | | Deposit date: | 2013-12-10 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of the NaV1.5 cytoplasmic domain by calmodulin.
Nat Commun, 5, 2014
|
|
5V68
 
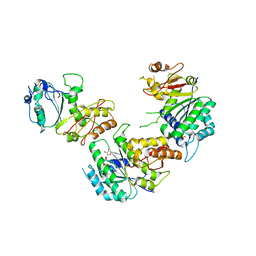 | | Crystal structure of cell division protein FtsZ from Mycobacterium tuberculosis bounded via the T9 loop | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Lazo, E.O, Ojima, I, Chowdhury, S.R, Awasthi, D, Jakoncic, J. | | Deposit date: | 2017-03-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Novel T9 loop conformation of filamenting temperature-sensitive mutant Z from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4X3T
 
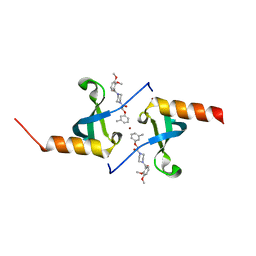 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with MS37452 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(2,3-dimethoxybenzoyl)piperazin-1-yl]-2-(3-methylphenoxy)ethanone, Chromobox protein homolog 7, ... | | Authors: | Ren, C, Jakoncic, J, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
3DWH
 
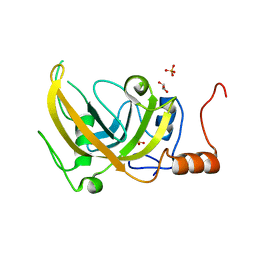 | |
3T6R
 
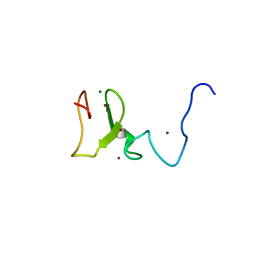 | | Structure of UHRF1 in complex with unmodified H3 N-terminal tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.1t N-terminal peptide, MAGNESIUM ION, ... | | Authors: | Xie, S, Jakoncic, J, Qian, C.M. | | Deposit date: | 2011-07-29 | | Release date: | 2011-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail
J.Mol.Biol., 415, 2012
|
|
4XDX
 
 | |
1XT5
 
 | | Crystal Structure of VCBP3, domain 1, from Branchiostoma floridae | | Descriptor: | SULFATE ION, variable region-containing chitin-binding protein 3 | | Authors: | Hernandez Prada, J.A, Haire, R.N, Cannon, J.P, Allaire, M, Jakoncic, J, Stojanoff, V, Litman, G.W, Ostrov, D.A. | | Deposit date: | 2004-10-21 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus.
Nat.Immunol., 7, 2006
|
|
8DCT
 
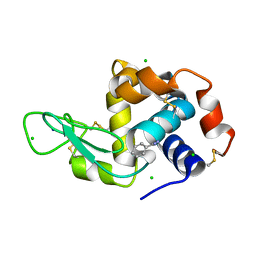 | | Lysozyme cluster 3 dual apo structure | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Soares, A.S, Yamada, Y, Jakoncic, J, Schneider, D.K, Bernstein, H.J. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serial crystallography with multi-stage merging of thousands of images.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8DCW
 
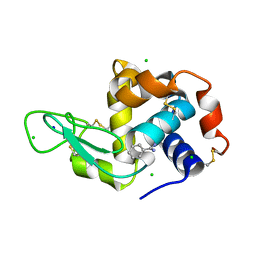 | | Lysozyme cluster 0062 (NAG and benzamidine ligands) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Soares, A.S, Yamada, Y, Jakoncic, J, Schneider, D.K, Bernstein, H.J. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serial crystallography with multi-stage merging of thousands of images.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8DCU
 
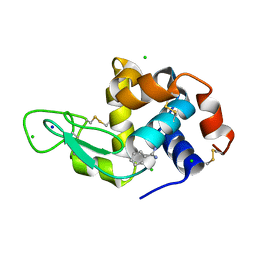 | | Lysozyme cluster 0028 (benzamidine ligand) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Soares, A.S, Yamada, Y, Jakoncic, J, Schneider, D.K, Bernstein, H.J. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serial crystallography with multi-stage merging of thousands of images.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8DCV
 
 | | Lysozyme cluster 0043, NAG ligand | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Soares, A.S, Yamada, Y, Jakoncic, J, Schneider, D.K, Bernstein, H.J. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serial crystallography with multi-stage merging of thousands of images.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6BSC
 
 | |
6BSB
 
 | |
4Z7A
 
 | | Structural and biochemical characterization of a non-functionally redundant M. tuberculosis (3,3) L,D-Transpeptidase, LdtMt5. | | Descriptor: | ACETYL GROUP, DI(HYDROXYETHYL)ETHER, Mycobacterium tuberculosis (3,3)L,D-Transpeptidase type 5, ... | | Authors: | Basta, L, Ghosh, A, Pan, Y, Jakoncic, J, Lloyd, E, Townsend, G, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-04-06 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Loss of a Functionally and Structurally Distinct ld-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
5AJL
 
 | | Sdsa sulfatase tetragonal | | Descriptor: | ALKYL SULFATASE, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncic, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
