6QZH
 
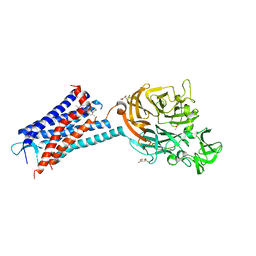 | | Structure of the human CC Chemokine Receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 and the insertion protein Sialidase NanA | | Descriptor: | 3-[[4-[[(1~{R})-2,2-dimethyl-1-(5-methylfuran-2-yl)propyl]amino]-1,1-bis(oxidanylidene)-1,2,5-thiadiazol-3-yl]amino]-~{N},~{N},6-trimethyl-2-oxidanyl-benzamide, C-C chemokine receptor type 7,Sialidase A,C-C chemokine receptor type 7, D(-)-TARTARIC ACID, ... | | Authors: | Jaeger, K, Bruenle, S, Weinert, T, Guba, W, Muehle, J, Miyazaki, T, Weber, M, Furrer, A, Haenggi, N, Tetaz, T, Huang, C.Y, Mattle, D, Vonach, J.M, Gast, A, Kuglstatter, A, Rudolph, M.G, Nogly, P, Benz, J, Dawson, R.J.P, Standfuss, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7.
Cell, 178, 2019
|
|
5OVM
 
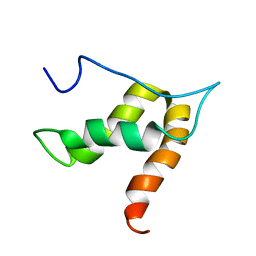 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2017-08-29 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
7QX3
 
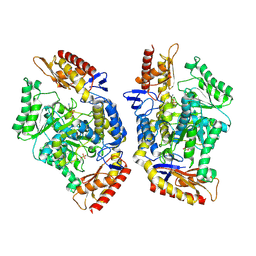 | | Structure of the transaminase TR2E2 with EOS | | Descriptor: | 2-azanylethyl hydrogen sulfate, Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7QX0
 
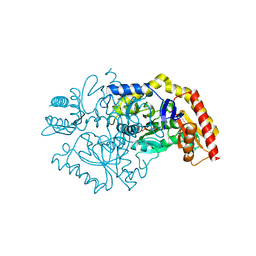 | | Transaminase Structure of Plurienzyme (Tr2E2) in complex with PLP | | Descriptor: | Aminotransferase TR2, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-26 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
3DCQ
 
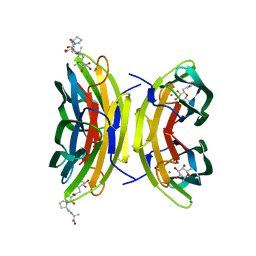 | | LECB (PA-LII) in complex with the synthetic ligand 2G0 | | Descriptor: | (2S)-1-[(2S)-6-amino-2-({[(2S,3S,4R,5S,6S)-3,4,5-trihydroxy-6-methyltetrahydro-2H-pyran-2-yl]acetyl}amino)hexanoyl]-N-[(1S)-1-carbamoyl-3-methylbutyl]pyrrolidine-2-carboxamide, CALCIUM ION, Fucose-binding lectin PA-IIL | | Authors: | Johansson, E.M, Crusz, S.A, Kolomiets, E, Buts, L, Kadam, R.U, Cacciarini, M, Bartels, K.M, Diggle, S.P, Camara, M, Williams, P, Loris, R, Nativi, C, Rosenau, F, Jaeger, K.E, Darbre, T, Reymond, J.L. | | Deposit date: | 2008-06-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition and dispersion of Pseudomonas aeruginosa biofilms by glycopeptide dendrimers targeting the fucose-specific lectin LecB.
Chem.Biol., 15, 2008
|
|
2YOM
 
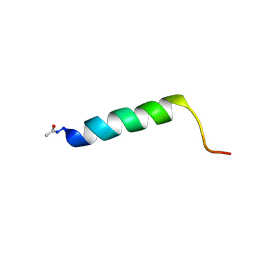 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Lecher, J, Hartmann, R, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
2YON
 
 | | Solution NMR structure of the C-terminal extension of two bacterial light, oxygen, voltage (LOV) photoreceptor proteins from Pseudomonas putida | | Descriptor: | SENSORY BOX PROTEIN | | Authors: | Rani, R, Hartmann, R, Lecher, J, Krauss, U, Jaeger, K, Willbold, D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Conservation of Dark Recovery Kinetic Parameters and Structural Features in the Pseudomonadaceae "Short" Light, Oxygen, Voltage (Lov) Protein Family: Implications for the Design of Lov-Based Optogenetic Tools.
Biochemistry, 52, 2013
|
|
4X31
 
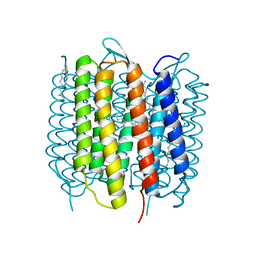 | | Room temperature structure of bacteriorhodopsin from lipidic cubic phase obtained with serial millisecond crystallography using synchrotron radiation | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, James, D, Wang, D, White, T, Zatsepin, N, Shilova, A, Nelson, G, Liu, H, Johansson, L, Heymann, M, Jaeger, K, Metz, M, Wickstrand, C, Wu, W, Baath, P, Berntsen, P, Oberthuer, D, Panneels, V, Cherezov, V, Chapman, H, Spence, J, Schertler, G, Neutze, R, Moraes, I, Burghammer, M, Standfuss, J, Weierstall, U. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipidic cubic phase serial millisecond crystallography using synchrotron radiation.
Iucrj, 2, 2015
|
|
4FBL
 
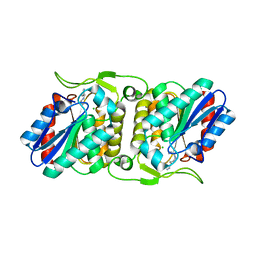 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
4FBM
 
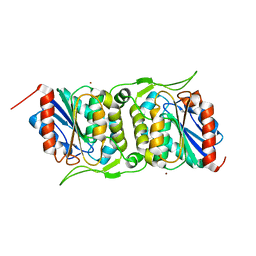 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
1I6W
 
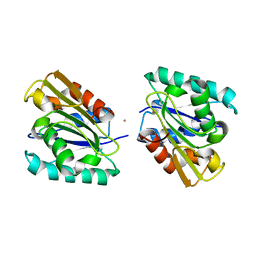 | | THE CRYSTAL STRUCTURE OF BACILLUS SUBTILIS LIPASE: A MINIMAL ALPHA/BETA HYDROLASE ENZYME | | Descriptor: | CADMIUM ION, LIPASE A | | Authors: | van Pouderoyen, G, Eggert, T, Jaeger, K.-E, Dijkstra, B.W. | | Deposit date: | 2001-03-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of Bacillus subtilis lipase: a minimal alpha/beta hydrolase fold enzyme.
J.Mol.Biol., 309, 2001
|
|
4KUO
 
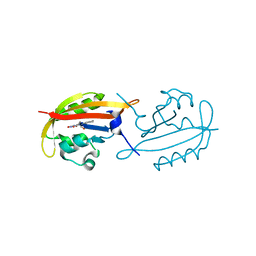 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Photoexcited state) | | Descriptor: | RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
6Z68
 
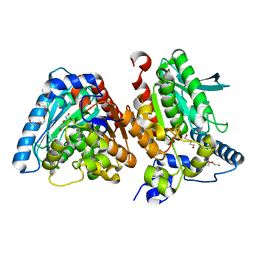 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | Acetyl esterase/lipase, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
6HRG
 
 | | Structure of Igni18, a novel metallo hydrolase from the hyperthermophilic archaeon Ignicoccus hospitalis KIN4/I | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, UPF0173 metal-dependent hydrolase Igni_1254, ... | | Authors: | Smits, S.H, Streit, W.R, Jaeger, K.E, Hoeppner, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A promiscuous ancestral enzyme ́s structure unveils protein variable regions of the highly diverse metallo-beta-lactamase family.
Commun Biol, 4, 2021
|
|
6Z69
 
 | | A novel metagenomic alpha/beta-fold esterase | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Acetyl esterase/lipase, MAGNESIUM ION, ... | | Authors: | Bollinger, A, Thies, S, Hoeppner, A, Kobus, S, Jaeger, K.-E, Smits, S.H.J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of a novel family IV esterase in free and substrate-bound form.
Febs J., 288, 2021
|
|
4JGG
 
 | |
4KUK
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
6SCD
 
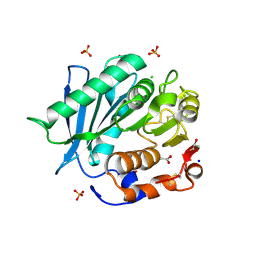 | | Polyester hydrolase PE-H Y250S mutant of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
6SBN
 
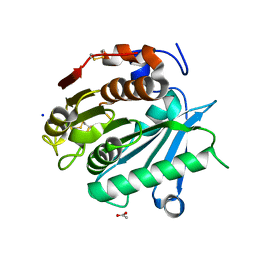 | | Polyester hydrolase PE-H of Pseudomonas aestusnigri | | Descriptor: | ACETATE ION, SODIUM ION, polyester hydrolase | | Authors: | Bollinger, A, Thies, S, Kobus, S, Hoeppner, A, Smits, S.H.J, Jaeger, K.-E. | | Deposit date: | 2019-07-22 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | A Novel Polyester Hydrolase From the Marine BacteriumPseudomonas aestusnigri -Structural and Functional Insights.
Front Microbiol, 11, 2020
|
|
7QYF
 
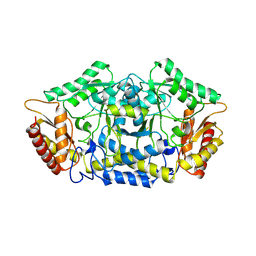 | | Structure of the transaminase PluriZyme variant (TR2E2) | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
7QYG
 
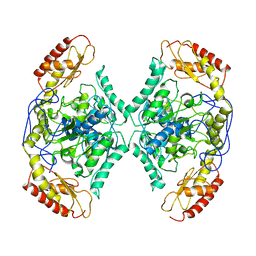 | | Structure of the transaminase TR2 | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
6GSF
 
 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2018-06-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
6G7L
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 8.3 ms state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7H
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: resting state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
6G7I
 
 | | Retinal isomerization in bacteriorhodopsin revealed by a femtosecond X-ray laser: 49-406 fs state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, ... | | Authors: | Nogly, P, Weinert, T, James, D, Cabajo, S, Ozerov, D, Furrer, A, Gashi, D, Borin, V, Skopintsev, P, Jaeger, K, Nass, K, Bath, P, Bosman, R, Koglin, J, Seaberg, M, Lane, T, Kekilli, D, Bruenle, S, Tanaka, T, Wu, W, Milne, C, White, T, Barty, A, Weierstall, U, Panneels, V, Nango, E, Iwata, S, Hunter, M, Schapiro, I, Schertler, G, Neutze, R, Standfuss, J. | | Deposit date: | 2018-04-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser.
Science, 361, 2018
|
|
