5DVL
 
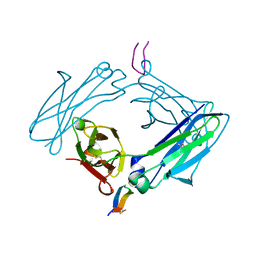 | | Fc Design 20.8.37 A chain homodimer Y349S/T366M/K370Y/K409V | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-30 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
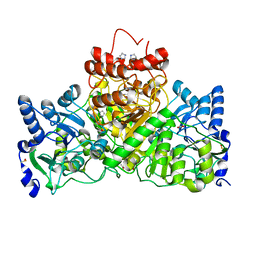
5DW4
 
 | |
5E54
 
 | | Two apo structures of the adenine riboswitch aptamer domain determined using an X-ray free electron laser | | Descriptor: | MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5DJY
 
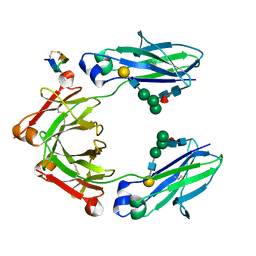 | | Fc Heterodimer Design 20.8 Y349S/T366V/K370Y/K409V + E357D/S364Q/Y407A | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-02 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
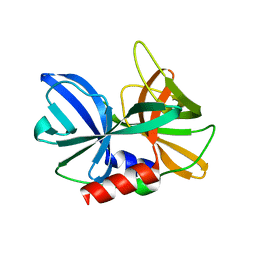
2J92
 
 | |
4N23
 
 | | Crystal structure of the GP2 Core Domain from the California Academy of Science Virus, monoclinic symmetry | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GP2 Ectodomain | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Dai, Z, Toro, R, Lai, J.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species.
J.Mol.Biol., 426, 2014
|
|
4IMV
 
 | |
4IGB
 
 | | Crystal structure of the N-terminal domain of the Streptococcus gordonii adhesin Sgo0707 | | Descriptor: | ACETATE ION, GLYCEROL, LPXTG cell wall surface protein, ... | | Authors: | Nylander, A, Svensater, G, Senadheera, D.B, Cvitkovitch, D.G, Davies, J.R, Persson, K. | | Deposit date: | 2012-12-17 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Functional Analysis of the N-terminal Domain of the Streptococcus gordonii Adhesin Sgo0707
Plos One, 8, 2013
|
|
4IBM
 
 | | Crystal structure of insulin receptor kinase domain in complex with an inhibitor Irfin-1 | | Descriptor: | 5-(2-phenylpyrazolo[1,5-a]pyridin-3-yl)-3H-pyrazolo[3,4-c]pyridazin-3-one, Insulin receptor | | Authors: | Wu, J, Anastassiadis, T, Duong-Ly, K.C, Peterson, J.R. | | Deposit date: | 2012-12-08 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A highly selective dual insulin receptor (IR)/insulin-like growth factor 1 receptor (IGF-1R) inhibitor derived from an extracellular signal-regulated kinase (ERK) inhibitor.
J.Biol.Chem., 288, 2013
|
|
4MYY
 
 | |
4MYZ
 
 | |
4JIU
 
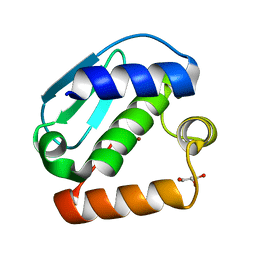 | | Crystal structure of the metallopeptidase zymogen of Pyrococcus abyssi abylysin | | Descriptor: | GLYCEROL, Proabylysin, ZINC ION | | Authors: | Lopez-Pelegrin, M, Cerda-Costa, N, Martinez-Jimenez, F, Cintas-Pedrola, A, Canals, A, Peinado, J.R, Marti-Renom, M.A, Lopez-Otin, C, Arolas, J.L, Gomis-Ruth, F.X. | | Deposit date: | 2013-03-07 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A novel family of soluble minimal scaffolds provides structural insight into the catalytic domains of integral membrane metallopeptidases
J.Biol.Chem., 288, 2013
|
|
4IL6
 
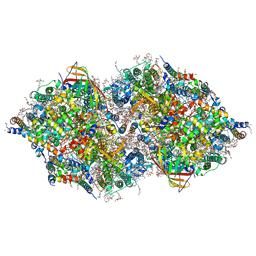 | | Structure of Sr-substituted photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Koua, F.H.M, Umena, Y, Kawakami, K, Kamiya, N, Shen, J.R. | | Deposit date: | 2012-12-29 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Sr-substituted photosystem II at 2.1 A resolution and its implications in the mechanism of water oxidation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IPE
 
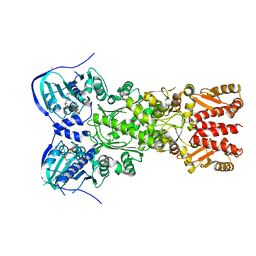 | | Crystal structure of mitochondrial Hsp90 (TRAP1) with AMPPNP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
4JLT
 
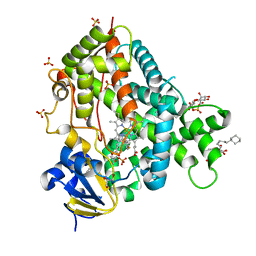 | | Crystal structure of P450 2B4(H226Y) in complex with paroxetine | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, GLYCEROL, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2013-03-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A Structural Snapshot of CYP2B4 in Complex with Paroxetine Provides Insights into Ligand Binding and Clusters of Conformational States.
J.Pharmacol.Exp.Ther., 346, 2013
|
|
2FLC
 
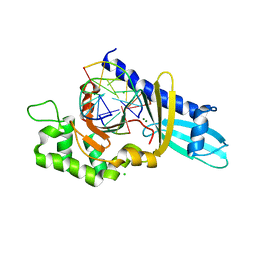 | |
4MT1
 
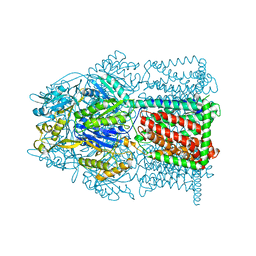 | |
4G4H
 
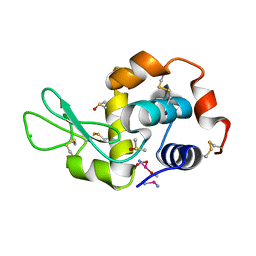 | | 100K X-ray diffraction study of carboplatin binding to HEWL in DMSO media after 13 months of crystal storage | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Schreurs, A.M.M, Kroon-Batenburg, L.M.J, Helliwell, J.R. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Room-temperature X-ray diffraction studies of cisplatin and carboplatin binding to His15 of HEWL after prolonged chemical exposure.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4IQN
 
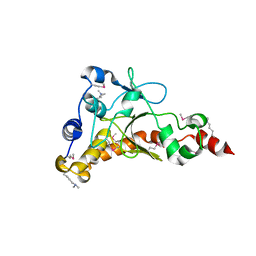 | | Crystal structure of uncharacterized protein from Salmonella enterica subsp. enterica serovar typhimurium str. 14028s | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative cytoplasmic protein, TETRAETHYLENE GLYCOL | | Authors: | Chang, C, Hatzos-Skintges, C, Adkins, J.N, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of uncharacterized protein from salmonella enterica subsp. enterica serovar typhimurium str. 14028s
To be Published
|
|
4K9J
 
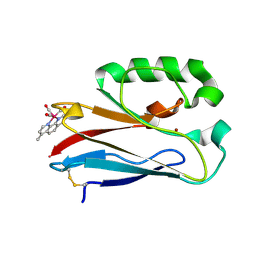 | | Structure of Re(CO)3(4,7-dimethyl-phen)(Thr126His)(Lys122Trp)(His83Glu)(Trp48Phe)(Tyr72Phe)(Tyr108Phe)AzCu(II), a Rhenium modified Azurin mutant | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Williamson, H.R, Blanco-Rodriguez, A.M, Sokolova, L, Nikolovski, P, Kaiser, J.T, Towrie, M, Clark, I.P, Vlcek Jr, A, Winkler, J.R, Gray, H.B. | | Deposit date: | 2013-04-20 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryptophan-accelerated electron flow across a protein-protein interface.
J.Am.Chem.Soc., 135, 2013
|
|
4N4P
 
 | | Crystal Structure of N-acetylneuraminate lyase from Mycoplasma synoviae, crystal form I | | Descriptor: | Acylneuraminate lyase, CHLORIDE ION | | Authors: | Georgescauld, F, Popova, K, Gupta, A.J, Bracher, A, Engen, J.R, Hayer-Hartl, M, Hartl, F.U. | | Deposit date: | 2013-10-08 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding.
Cell(Cambridge,Mass.), 157, 2014
|
|
2JBK
 
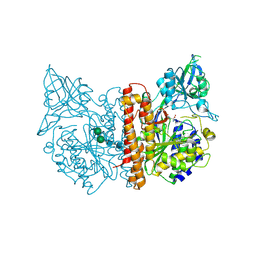 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with quisqualic acid (quisqualate, alpha-amino-3,5-dioxo-1,2,4- oxadiazolidine-2-propanoic acid) | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Henning, K, Hilgenfeld, R. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JBJ
 
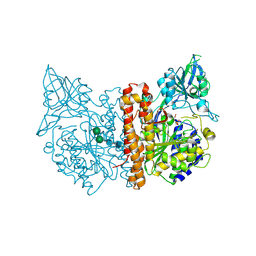 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with 2-PMPA (2-phosphonoMethyl-pentanedioic acid) | | Descriptor: | (2S)-2-(PHOSPHONOMETHYL)PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Henning, K, Hilgenfeld, R. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4IUW
 
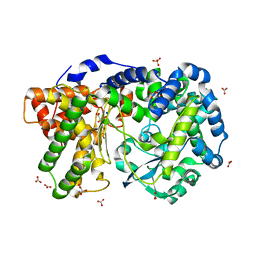 | | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20) | | Descriptor: | CARBONATE ION, CITRIC ACID, Neutral endopeptidase, ... | | Authors: | Anderson, B.F, Knapp, K.M, Holland, R, Norris, G.E, Christensson, C, Bratt, H, Collins, L.J, Coolbear, T, Lubbers, M.W, Toole, P.W.O, Reid, J.R, Jameson, G.B. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20)
TO BE PUBLISHED
|
|
4J0B
 
 | | Structure of mitochondrial Hsp90 (TRAP1) with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, COBALT (II) ION, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-30 | | Release date: | 2014-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
