2ZTC
 
 | | MtRuvA Form II | | Descriptor: | GLYCEROL, Holliday junction ATP-dependent DNA helicase ruvA | | Authors: | Prabu, J.R, Thamotharan, S, Khanduja, J.S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-10-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic and modelling studies on Mycobacterium tuberculosis RuvA Additional role of RuvB-binding domain and inter species variability
Biochim.Biophys.Acta, 1794, 2009
|
|
2ZR0
 
 | | MSRECA-Q196E mutant | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein recA | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-22 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3B7Y
 
 | | Crystal structure of the C2 Domain of the E3 Ubiquitin-Protein Ligase NEDD4 | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase NEDD4 | | Authors: | Walker, J.R, Ruzanov, M, Butler-Cole, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C2 Domain of the Human E3 Ubiquitin-Protein Ligase NEDD4.
To be Published
|
|
3BAI
 
 | | Human Pancreatic Alpha Amylase with Bound Nitrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NITRATE ION, ... | | Authors: | Fredriksen, J.R, Maurus, R, Brayer, G.D. | | Deposit date: | 2007-11-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
3B6Q
 
 | |
3B6W
 
 | |
3BI7
 
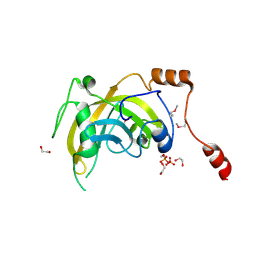 | | Crystal structure of the SRA domain of E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, SULFATE ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1.
Nature, 455, 2008
|
|
3BU8
 
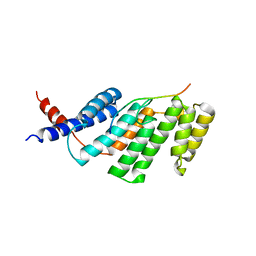 | | Crystal Structure of TRF2 TRFH domain and TIN2 peptide complex | | Descriptor: | TERF1-interacting nuclear factor 2, Telomeric repeat-binding factor 2 | | Authors: | Chen, Y, Yang, Y, van Overbeek, M, Donigian, J.R, Baciu, P, de Lange, T, Lei, M. | | Deposit date: | 2008-01-02 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins.
Science, 319, 2008
|
|
3BRB
 
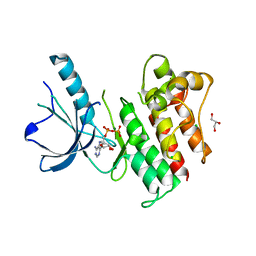 | | Crystal structure of catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
3C8X
 
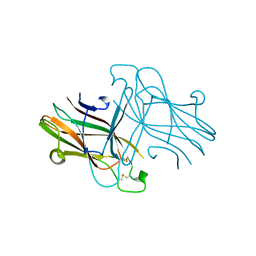 | | Crystal structure of the ligand binding domain of human Ephrin A2 (Epha2) receptor protein kinase | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8Q7R
 
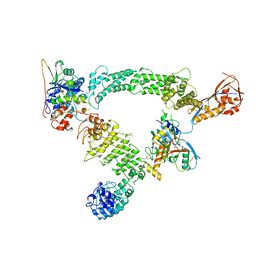 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | Descriptor: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-01 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4RA0
 
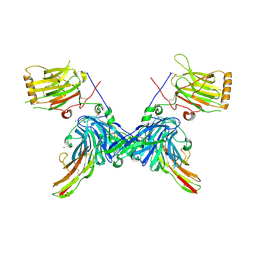 | | An engineered Axl 'decoy receptor' effectively silences the Gas6-Axl signaling axis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kariolis, M.S, Kapur, S, Mathews, I.I, Cochran, J.R. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | An engineered Axl 'decoy receptor' effectively silences the Gas6-Axl signaling axis.
Nat.Chem.Biol., 10, 2014
|
|
4R0C
 
 | | Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology | | Descriptor: | AbgT putative transporter family, DODECYL-BETA-D-MALTOSIDE, SODIUM ION, ... | | Authors: | Su, C.-C, Bolla, J.R, Yu, E.W. | | Deposit date: | 2014-07-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology.
Nat Commun, 6, 2015
|
|
4R9F
 
 | | CpMnBP1 with Mannobiose Bound | | Descriptor: | MBP1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Chekan, J.R, Agarwal, V, Nair, S.K. | | Deposit date: | 2014-09-04 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Basis for Mannan Utilization by Caldanaerobius polysaccharolyticus Strain ATCC BAA-17.
J.Biol.Chem., 289, 2014
|
|
4R9G
 
 | | CpMnBP1 with Mannotriose Bound | | Descriptor: | MBP1, ZINC ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Chekan, J.R, Agarwal, V, Nair, S.K. | | Deposit date: | 2014-09-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Biochemical Basis for Mannan Utilization by Caldanaerobius polysaccharolyticus Strain ATCC BAA-17.
J.Biol.Chem., 289, 2014
|
|
4RFQ
 
 | | Human Methyltransferase-Like 18 | | Descriptor: | Histidine protein methyltransferase 1 homolog, S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Ravichandran, M, Li, Y, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Hong, B.S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human Methyltransferase-Like 18
TO BE PUBLISHED
|
|
4QPW
 
 | | BiXyn10A CBM1 with Xylohexaose Bound | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2014-06-25 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | Descriptor: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RE2
 
 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | Descriptor: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QCU
 
 | | Lysozyme covalently bound to fac-[Re(CO)3-imidazole] complex, incubated for 38 weeks. | | Descriptor: | BROMIDE ION, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Jacobs, F.J.F, Brink, A, Helliwell, J.R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Time-series analysis of rhenium(I) organometallic covalent binding to a model protein for drug development.
Iucrj, 11, 2024
|
|
4QPP
 
 | | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with compound DS-421 (2-{4-[3-CHLORO-2-(2-METHOXYPHENYL)-1H-INDOL-5-YL]PIPERIDIN-1-YL}-N-METHYLETHANAMINE | | Descriptor: | 2-{4-[3-chloro-2-(2-methoxyphenyl)-1H-indol-5-yl]piperidin-1-yl}-N-methylethanamine, POLY-UNK, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, Smil, D, Walker, J.R, He, H, Eram, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Crystal Structure of Human HMT1
hnRNP methyltransferase-like protein 6 in complex with compound DS-421
To be Published
|
|
4R6C
 
 | |
4RE4
 
 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Beta-mannosidase/beta-glucosidase, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
