7TT1
 
 | | BamABCDE bound to substrate EspP class 4 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT5
 
 | | BamABCDE bound to substrate EspP in the open-sheet EspP state | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamC, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7TT2
 
 | | BamABCDE bound to substrate EspP class 3 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Serine protease EspP chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Doyle, M.T, Jimah, J.R, Dowdy, T, Ohlemacher, S.I, Larion, M, Hinshaw, J.E, Bernstein, H.D. | | Deposit date: | 2022-01-31 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding.
Cell, 185, 2022
|
|
7VD5
 
 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nagao, R, Kato, K, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2021-09-06 | | Release date: | 2022-03-02 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for different types of hetero-tetrameric light-harvesting complexes in a diatom PSII-FCPII supercomplex
Nat Commun, 13, 2022
|
|
4FF9
 
 | | Crystal Structure of cysteinylated WT SOD1. | | Descriptor: | COPPER (II) ION, CYSTEINE, Superoxide dismutase [Cu-Zn], ... | | Authors: | Auclair, J.R, Brodkin, H.R, D'Aquino, J.A, Ringe, D, Petsko, G.A, Agar, J.N. | | Deposit date: | 2012-05-31 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5003 Å) | | Cite: | Structural consequences of cysteinylation of cu/zn-superoxide dismutase.
Biochemistry, 52, 2013
|
|
4FNJ
 
 | |
4FHT
 
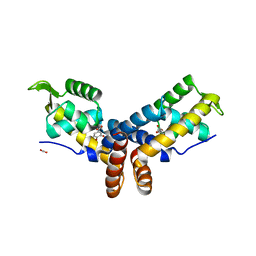 | | Crystal Structure of the PcaV transcriptional regulator from Streptomyces coelicolor in complex with its natural ligand | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, PcaV transcriptional regulator | | Authors: | Brown, B.L, Davis, J.R, Sello, J.K, Page, R. | | Deposit date: | 2012-06-06 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Study of PcaV from Streptomyces coelicolor yields new insights into ligand-responsive MarR family transcription factors.
Nucleic Acids Res., 41, 2013
|
|
4G4H
 
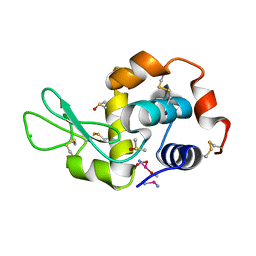 | | 100K X-ray diffraction study of carboplatin binding to HEWL in DMSO media after 13 months of crystal storage | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Schreurs, A.M.M, Kroon-Batenburg, L.M.J, Helliwell, J.R. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Room-temperature X-ray diffraction studies of cisplatin and carboplatin binding to His15 of HEWL after prolonged chemical exposure.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4G9Y
 
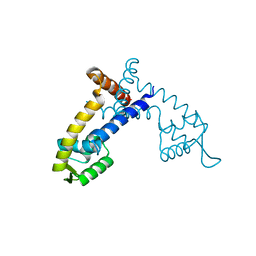 | | Crystal Structure of the PcaV transcriptional regulator from Streptomyces coelicolor | | Descriptor: | GLYCEROL, PcaV transcriptional regulator | | Authors: | Brown, B.L, Davis, J.R, Sello, J.K, Page, R. | | Deposit date: | 2012-07-24 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Study of PcaV from Streptomyces coelicolor yields new insights into ligand-responsive MarR family transcription factors.
Nucleic Acids Res., 41, 2013
|
|
4G49
 
 | | Room temperature X-ray diffraction of cisplatin binding to HEWL in aqueous media after 15 months of crystal storage | | Descriptor: | ACETATE ION, CHLORIDE ION, Cisplatin, ... | | Authors: | Tanley, S.W.M, Schreurs, A.M.M, Kroon-Batenburg, L.M.J, Helliwell, J.R. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room-temperature X-ray diffraction studies of cisplatin and carboplatin binding to His15 of HEWL after prolonged chemical exposure.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4GCE
 
 | |
5MIL
 
 | | Pirating conserved phage mechanisms promotes promiscuous staphylococcal pathogenicity islands transfer. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase family protein, MAGNESIUM ION | | Authors: | Donderis, J, Marina, A, Penades, J.R. | | Deposit date: | 2016-11-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pirating conserved phage mechanisms promotes promiscuous staphylococcal pathogenicity island transfer.
Elife, 6, 2017
|
|
4V51
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with mRNA, tRNA and paromomycin | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Selmer, M, Dunham, C.M, Murphy, F.V, Weixlbaumer, A, Petry, S, Weir, J.R, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2006-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the 70S ribosome complexed with mRNA and tRNA.
Science, 313, 2006
|
|
4V8Y
 
 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
4W8R
 
 | |
4UY3
 
 | | Cytoplasmic domain of bacterial cell division protein ezra | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
4UWJ
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a capped pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-N-(difluoromethyl)-4-[3-(piperidin-4-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
4UWI
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-4-[3-(4-methylpiperazin-1-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
1CK2
 
 | |
1CEG
 
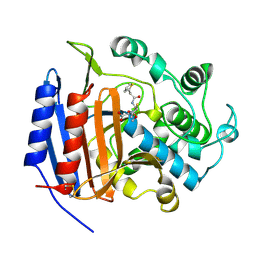 | | CEPHALOTHIN COMPLEXED WITH DD-PEPTIDASE | | Descriptor: | CEPHALOTHIN GROUP, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE TRANSPEPTIDASE | | Authors: | Knox, J.R, Kuzin, A.P. | | Deposit date: | 1995-01-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of cephalothin and cefotaxime to D-ala-D-ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum beta-lactamases.
Biochemistry, 34, 1995
|
|
1CN7
 
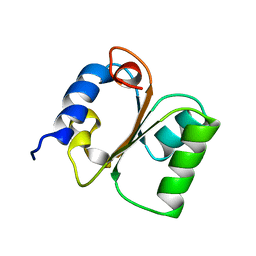 | | Yeast ribosomal protein L30 | | Descriptor: | 60S RIBOSOMAL PROTEIN L30E | | Authors: | Mao, H, Willamson, J.R. | | Deposit date: | 1999-05-26 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Local folding coupled to RNA binding in the yeast ribosomal protein L30.
J.Mol.Biol., 292, 1999
|
|
1CEF
 
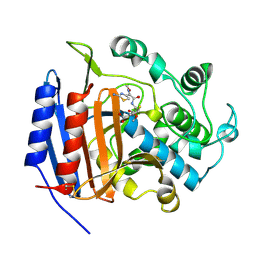 | | CEFOTAXIME COMPLEXED WITH THE STREPTOMYCES R61 DD-PEPTIDASE | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Knox, J.R, Kuzin, A.P. | | Deposit date: | 1995-01-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Binding of cephalothin and cefotaxime to D-ala-D-ala-peptidase reveals a functional basis of a natural mutation in a low-affinity penicillin-binding protein and in extended-spectrum beta-lactamases.
Biochemistry, 34, 1995
|
|
1DDN
 
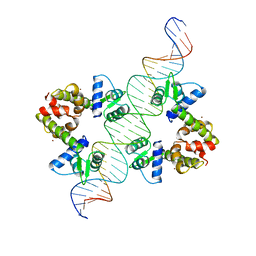 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT)/TOX DNA OPERATOR COMPLEX | | Descriptor: | 33 BASE DNA CONTAINING TOXIN OPERATOR, DIPHTHERIA TOX REPRESSOR, NICKEL (II) ION | | Authors: | White, A, Ding, X, Vanderspek, J.C, Murphy, J.R, Ringe, D. | | Deposit date: | 1998-06-23 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex.
Nature, 394, 1998
|
|
5L3H
 
 | |
7SSX
 
 | | Structure of human Kv1.3 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
