3IW6
 
 | | Human p38 MAP Kinase in Complex with a Benzylpiperazin-Pyrrol | | Descriptor: | Mitogen-activated protein kinase 14, ethyl 4-[(4-benzylpiperazin-1-yl)carbonyl]-1-ethyl-3,5-dimethyl-1H-pyrrole-2-carboxylate, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
2SHK
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF SHIKIMATE KINASE FROM ERWINIA CHRYSANTHEMI COMPLEXED WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SHIKIMATE KINASE | | Authors: | Krell, T, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallization and preliminary X-ray crystallographic analysis of shikimate kinase from Erwinia chrysanthemi.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
7QFQ
 
 | | Cryo-EM structure of Botulinum neurotoxin serotype B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
1DVD
 
 | | SOLUTION NMR STRUCTURE OF HUMAN STEFIN A AT PH 5.5 AND 308K, NMR, 17 STRUCTURES | | Descriptor: | STEFIN A | | Authors: | Martin, J.R, Craven, C.J, Jerala, R, Kroon-Zitko, L, Zerovnik, E, Turk, V, Waltho, J.P. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of human stefin A.
J.Mol.Biol., 246, 1995
|
|
6JEO
 
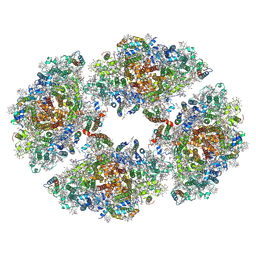 | | Structure of PSI tetramer from Anabaena | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-02-06 | | Release date: | 2019-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy.
Nat Commun, 10, 2019
|
|
6JLN
 
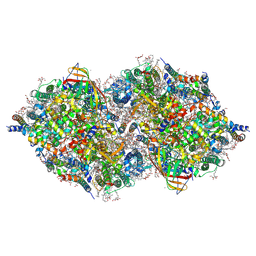 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
3ICS
 
 | |
3IDY
 
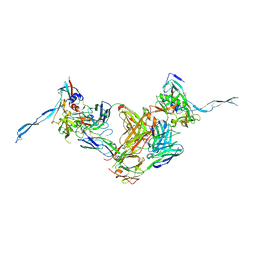 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab b13 heavy chain, Fab b13 light chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IW5
 
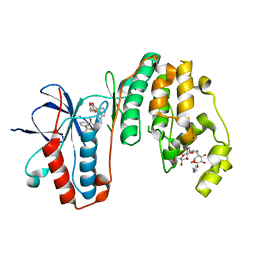 | | Human p38 MAP Kinase in Complex with an Indole Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-(3-{[2-(2,3-dihydro-1,4-benzodioxin-6-ylamino)-2-oxoethyl]sulfanyl}-1H-indol-1-yl)ethyl]-3-(trifluoromethyl)benzamide, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
1DQU
 
 | | CRYSTAL STRUCTURE OF THE ISOCITRATE LYASE FROM ASPERGILLUS NIDULANS | | Descriptor: | ISOCITRATE LYASE | | Authors: | Britton, K.L, Langridge, S.J, Baker, P.J, Weeradechapon, K, Sedelnikova, S.E, De Lucas, J.R, Rice, D.W, Turner, G. | | Deposit date: | 2000-01-05 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure and active site location of isocitrate lyase from the fungus Aspergillus nidulans.
Structure Fold.Des., 8, 2000
|
|
6HGB
 
 | | Influenza A virus N6 neuraminidase native structure (Duck/England/56). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salinger, M.T, Hobbs, J.R, Murray, J.W, Laver, W.G, Kuhn, P, Garman, E.F. | | Deposit date: | 2018-08-23 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Structures of Viral Neuraminidase with Drugs Bound in the Active Site. (In preparation)
To Be Published
|
|
3IHP
 
 | | Covalent Ubiquitin-Usp5 Complex | | Descriptor: | CHLORIDE ION, ETHANAMINE, Ubiquitin, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Covalent Ubiquitin-Usp5 Complex
To be Published
|
|
3IYI
 
 | | P22 expanded head coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links | | Descriptor: | P22 coat protein in procapsid shells | | Authors: | Parent, K.N, Khayat, R, Tu, L.H, Suhanovsky, M.M, Cortines, J.R, Teschke, C.M, Johnson, J.E, Baker, T.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks
Structure, 18, 2010
|
|
6JLK
 
 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLM
 
 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
3ICR
 
 | | Crystal structure of oxidized Bacillus anthracis CoADR-RHD | | Descriptor: | COENZYME A, Coenzyme A-Disulfide Reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Wallen, J.R, Claiborne, A. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
Biochemistry, 48, 2009
|
|
8D1U
 
 | | E. coli beta-ketoacyl-[acyl carrier protein] synthase III (FabH) with an acetylated cysteine and in complex with oxa(dethia)-Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, CHLORIDE ION, oxa(dethia)-CoA | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-ketoacylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(dethia)CoA.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3IW8
 
 | | Structure of Inactive Human p38 MAP Kinase in Complex with a Thiazole-Urea | | Descriptor: | 1-{4-[(1S)-1-amino-2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}-3-(3-chloro-4-fluorophenyl)urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
8CXL
 
 | | Structure of NapH3, a vanadium-dependent haloperoxidase homolog catalyzing the stereospecific alpha-hydroxyketone rearrangement reaction in napyradiomycin biosynthesis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NapH3 | | Authors: | Chen, P.Y.-T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 61, 2022
|
|
3IBD
 
 | | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Gay, S.C, Sun, L, Talakad, J.C, Shah, M.B, Stout, D.C, Halpert, J.R. | | Deposit date: | 2009-07-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution.
Mol.Pharmacol., 77, 2010
|
|
3ICV
 
 | | Structural Consequences of a Circular Permutation on Lipase B from Candida Antartica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
3ICU
 
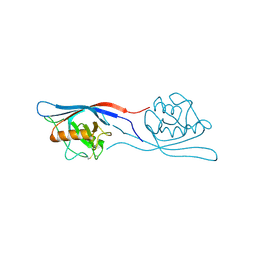 | | Protease-associated domain of the E3 ligase grail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, E3 ubiquitin-protein ligase RNF128 | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-18 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PA Domain of the E3 Ligase Grail
To be Published
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D3C
 
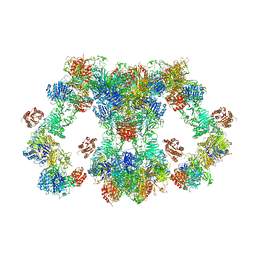 | | VWF tubule derived from monomeric D1-A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand factor | | Authors: | Anderson, J.R, Li, J, Springer, T.A, Brown, A. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of VWF tubules before and after concatemerization reveal a mechanism of disulfide bond exchange.
Blood, 140, 2022
|
|
