2O25
 
 | | Ubiquitin-Conjugating Enzyme E2-25 kDa Complexed With SUMO-1-Conjugating Enzyme UBC9 | | Descriptor: | SUMO-1-conjugating enzyme UBC9, Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel and Unexpected Complex Between the SUMO-1-Conjugating Enzyme UBC9 and the Ubiquitin-Conjugating Enzyme E2-25 kDa
To be Published
|
|
6WF6
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase | | Descriptor: | COBALT (II) ION, DI(HYDROXYETHYL)ETHER, Methylmalonyl-CoA epimerase | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
6WFH
 
 | | Streptomyces coelicolor methylmalonyl-CoA epimerase substrate complex | | Descriptor: | (3S,5R,9R,19E)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,19-tetrahydroxy-8,8,20-trimethyl-10,14-dioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphahenicos-19-en-21-oic acid 3,5-dioxide (non-preferred name), CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Stunkard, L.M, Benjamin, A.B, Bower, J.B, Huth, T.J, Lohman, J.R. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substrate Enolate Intermediate and Mimic Captured in the Active Site of Streptomyces coelicolor Methylmalonyl-CoA Epimerase.
Chembiochem, 23, 2022
|
|
7URJ
 
 | |
3MX0
 
 | | Crystal Structure of EphA2 ectodomain in complex with ephrin-A5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-A receptor 2, ... | | Authors: | Himanen, J.P, Yermekbayeva, L, Janes, P.W, Walker, J.R, Xu, K, Atapattu, L, Rajashankar, K.R, Mensinga, A, Lackmann, M, Nikolov, D.B, Dhe-Paganon, S. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1BLQ
 
 | | STRUCTURE AND INTERACTION SITE OF THE REGULATORY DOMAIN OF TROPONIN-C WHEN COMPLEXED WITH THE 96-148 REGION OF TROPONIN-I, NMR, 29 STRUCTURES | | Descriptor: | N-TROPONIN C | | Authors: | Mckay, R.T, Pearlstone, J.R, Corson, D.C, Gagne, S.M, Smillie, L.B, Sykes, B.D. | | Deposit date: | 1998-07-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and interaction site of the regulatory domain of troponin-C when complexed with the 96-148 region of troponin-I.
Biochemistry, 37, 1998
|
|
2OCB
 
 | | Crystal structure of human RAB9B in complex with a GTP analogue | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein Rab-9B, ... | | Authors: | Hong, B, Shen, L, Walker, J.R, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human RAB9B in complex with a GTP analogue
To be Published
|
|
1BKM
 
 | | COCRYSTAL STRUCTURE OF D-AMINO ACID SUBSTITUTED PHOSPHOPEPTIDE COMPLEX | | Descriptor: | PP60 V-SRC TYROSINE KINASE TRANSFORMING PROTEIN, [[O-PHOSPHONO-N-ACETYL-TYROSINYL]-GLUTAMYL-3[CYCLOHEXYLMETHYL]ALANINYL]-AMINE | | Authors: | Holland, D.R, Rubin, J.R. | | Deposit date: | 1997-05-02 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of a Novel Series of Nonpeptide Ligands that Bind to the Pp60Src Sh2 Domain
J.Am.Chem.Soc., 119, 1997
|
|
3MZK
 
 | | Sec13/Sec16 complex, S.cerevisiae | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC16 | | Authors: | Whittle, J.R, Schwartz, T.U. | | Deposit date: | 2010-05-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat.
J.Cell Biol., 190, 2010
|
|
6UKH
 
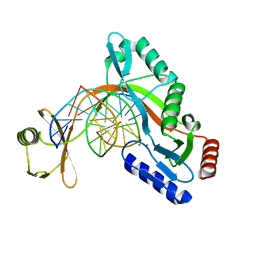 | | HhaI endonuclease in Complex with DNA in space group P41212 (pH 6.0) | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*GP*CP*GP*CP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*GP*CP*GP*CP*TP*TP*GP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2019-10-04 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure of HhaI endonuclease with cognate DNA at an atomic resolution of 1.0 angstrom.
Nucleic Acids Res., 48, 2020
|
|
1D43
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE UP | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1CVU
 
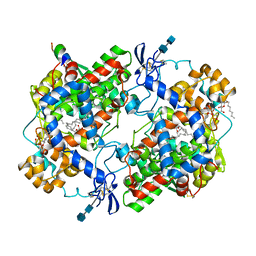 | | CRYSTAL STRUCTURE OF ARACHIDONIC ACID BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, ... | | Authors: | Kiefer, J.R, Pawlitz, J.L, Moreland, K.T, Stegeman, R.A, Gierse, J.K, Stevens, A.M, Goodwin, D.C, Rowlinson, S.W, Marnett, L.J, Stallings, W.C, Kurumbail, R.G. | | Deposit date: | 1999-08-24 | | Release date: | 2000-05-16 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the stereochemistry of the cyclooxygenase reaction.
Nature, 405, 2000
|
|
2VB0
 
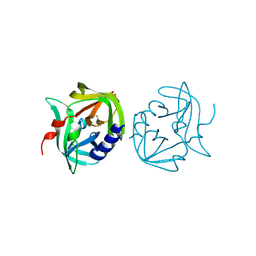 | | Crystal structure of coxsackievirus B3 proteinase 3C | | Descriptor: | CHLORIDE ION, POLYPROTEIN 3BCD | | Authors: | Anand, K, Mesters, J.R, Goerlach, R, Zell, R, Hilgenfeld, R. | | Deposit date: | 2007-09-05 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Coxsackie Virus B3 Proteinase 3C
To be Published
|
|
2O9C
 
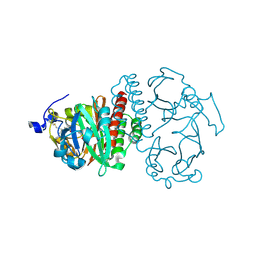 | | Crystal Structure of Bacteriophytochrome chromophore binding domain at 1.45 angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Wagner, J.R, Brunzelle, J.S, Vierstra, R.D, Forest, K.T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution structure of deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution.
J.Biol.Chem., 282, 2007
|
|
2OB0
 
 | | Human MAK3 homolog in complex with Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Human MAK3 homolog | | Authors: | Walker, J.R, Schuetz, S, Antoshenko, T, Wu, H, Bernstein, G, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-18 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human MAK3 homolog
To be Published
|
|
6WEA
 
 | | YTH domain of human YTHDC1 with a 10mer Oligo Containing N6mA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*GP*(6MA)P*CP*TP*TP*C)-3'), SODIUM ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
1D0I
 
 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR COMPLEXED WITH PHOSPHATE IONS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, TYPE II 3-DEHYDROQUINATE HYDRATASE | | Authors: | Roszak, A.W, Krell, T, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1999-09-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and mechanism of the type II dehydroquinase from Streptomyces coelicolor.
Structure, 10, 2002
|
|
1D49
 
 | | THE STRUCTURE OF A B-DNA DECAMER WITH A CENTRAL T-A STEP: C-G-A-T-T-A-A-T-C-G | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*AP*AP*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Grzeskowiak, K, Yanagi, K, Dickerson, R.E. | | Deposit date: | 1991-09-17 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G.
J.Mol.Biol., 225, 1992
|
|
2WNF
 
 | | Crystal Structure of a Mammalian Sialyltransferase in complex with Gal-beta-1-3GalNAc-ortho-nitrophenol | | Descriptor: | CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, HYDROXY(2-HYDROXYPHENYL)OXOAMMONIUM, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6WOK
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 6 | | Descriptor: | (1R,3R)-1-(2,6-difluoro-4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Labadie, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Discovery of GNE-149 as a Full Antagonist and Efficient Degrader of Estrogen Receptor alpha for ER+ Breast Cancer.
Acs Med.Chem.Lett., 11, 2020
|
|
3PA6
 
 | |
2WML
 
 | | Crystal Structure of a Mammalian Sialyltransferase | | Descriptor: | CHLORIDE ION, CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE -ALPHA-2,3-SIALYLTRANSFERASE, GLYCEROL | | Authors: | Rao, F.V, Rich, J.R, Raikic, B, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2009-07-01 | | Release date: | 2009-10-13 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight Into Mammalian Sialyltransferases.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3F5L
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-04 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
2PUQ
 
 | |
6WJR
 
 | |
