7VUS
 
 | | Crystal structure of AlleyCat9 with 5-nitro-benzotriazole | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1H-benzotriazole, AlleyCat, ... | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUR
 
 | | Crystal structure of AlleyCat9 with calcium but no inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AlleyCat, CALCIUM ION | | Authors: | Margheritis, E, Takahashi, K, Korendovych, I.V, Tame, J.R.H. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
7VUC
 
 | |
3CGE
 
 | |
7VUU
 
 | | Crystal structure of AlleyCat10 with inhibitor | | Descriptor: | 5-nitro-1H-benzotriazole, AlleyCat, CALCIUM ION | | Authors: | Tame, J.R.H, Korendovych, I.V, Margheritis, E, Takahashi, K. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
3MVD
 
 | | Crystal structure of the chromatin factor RCC1 in complex with the nucleosome core particle | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Makde, R.D, England, J.R, Yennawar, H.P, Tan, S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of RCC1 chromatin factor bound to the nucleosome core particle.
Nature, 467, 2010
|
|
8EHW
 
 | |
6VWT
 
 | |
2X61
 
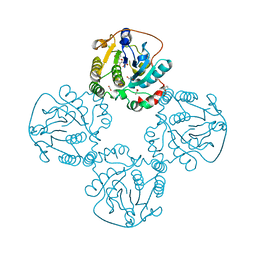 | | Crystal structure of the sialyltransferase CST-II in complex with trisaccharide acceptor and CMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Lee, H.J, Lairson, L.L, Rich, J.R, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2010-02-16 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Kinetic Analysis of Substrate Binding to the Sialyltransferase Cst-II from Campylobacter Jejuni.
J.Biol.Chem., 286, 2011
|
|
3MTN
 
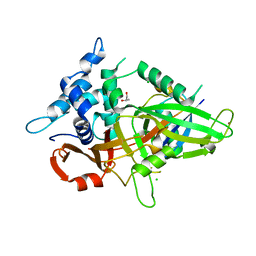 | | Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, UBIQUITIN VARIANT UBV.21.4, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
2UWL
 
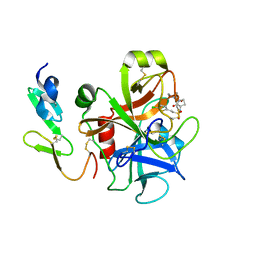 | | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHENESULFONAMIDE, COAGULATION FACTOR X | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P. | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2V58
 
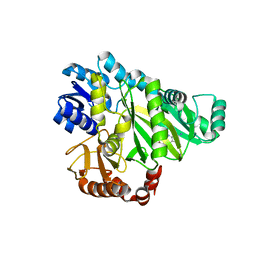 | | CRYSTAL STRUCTURE OF BIOTIN CARBOXYLASE FROM E.COLI IN COMPLEX WITH POTENT INHIBITOR 1 | | Descriptor: | 6-(2,6-dibromophenyl)pyrido[2,3-d]pyrimidine-2,7-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Class of Selective Antibacterials Derived from a Protein Kinase Inhibitor Pharmacophore.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3CGC
 
 | |
2ONI
 
 | | Catalytic Domain of the Human NEDD4-like E3 Ligase | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like protein, SODIUM ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-24 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NEDD4-like E3 Ubiquitin-protein Ligase
To be Published
|
|
2VJ1
 
 | | A Structural View of the Inactivation of the SARS-Coronavirus Main Proteinase by Benzotriazole Esters | | Descriptor: | 4-(DIMETHYLAMINO)BENZOIC ACID, BENZOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Verschueren, K.H.G, Pumpor, K, Anemueller, S, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural View of the Inactivation of the Sars Coronavirus Main Proteinase by Benzotriazole Esters.
Chem.Biol., 15, 2008
|
|
3CEG
 
 | | Crystal structure of the UBC domain of baculoviral IAP repeat-containing protein 6 | | Descriptor: | Baculoviral IAP repeat-containing protein 6 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-04-01 | | Last modified: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
2V6N
 
 | | Crystal structures of the SARS-coronavirus main proteinase inactivated by benzotriazole compounds | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(DIMETHYLAMINO)BENZOIC ACID, REPLICASE POLYPROTEIN 1AB, ... | | Authors: | Verschueren, K.H.G, Pumpor, K, Anemueller, S, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2007-07-19 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Structural View of the Inactivation of the Sars Coronavirus Main Proteinase by Benzotriazole Esters.
Chem.Biol., 15, 2008
|
|
2V5A
 
 | |
3CX2
 
 | | Crystal structure of the C1 domain of cardiac isoform of myosin binding protein-C at 1.3A | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Fisher, S.J, Helliwell, J.R, Khurshid, S, Govada, L, Redwood, C, Squire, J.M, Chayen, N.E. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An investigation into the protonation states of the C1 domain of cardiac myosin-binding protein C
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2OB4
 
 | | Human Ubiquitin-Conjugating Enzyme CDC34 | | Descriptor: | Ubiquitin-conjugating enzyme E2-32 kDa complementing | | Authors: | Neculai, D, Avvakumov, G.V, Xue, S, Walker, J.R, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Sicheri, F, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-18 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
3MZL
 
 | | Sec13/Sec31 edge element, loop deletion mutant | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Whittle, J.R, Schwartz, T.U. | | Deposit date: | 2010-05-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat.
J.Cell Biol., 190, 2010
|
|
6UHO
 
 | | Crystal Structure of C148 mGFP-cDNA-2 | | Descriptor: | C148 mGFP-cDNA-2, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
1CBI
 
 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN I | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN I | | Authors: | Thompson, J.R, Bratt, J.M, Banaszak, L.J. | | Deposit date: | 1995-07-12 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cellular retinoic acid binding protein I shows increased access to the binding cavity due to formation of an intermolecular beta-sheet.
J.Mol.Biol., 252, 1995
|
|
2WYG
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1R)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6WE9
 
 | | YTH domain of human YTHDC1 with 11mer ssDNA Containing N6mA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*CP*GP*GP*(6MA)P*CP*TP*CP*TP*G)-3'), GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
