3OA5
 
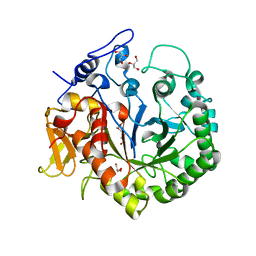 | | The structure of chi1, a chitinase from Yersinia entomophaga | | Descriptor: | Chi1, GLYCEROL, NONAETHYLENE GLYCOL | | Authors: | Busby, J.N, Lott, J.S, Hurst, M.R.H. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of Chi1 Chitinase from Yen-Tc: the multisubunit insecticidal ABC toxin complex of Yersinia entomophaga
J.Mol.Biol., 415, 2012
|
|
3QNR
 
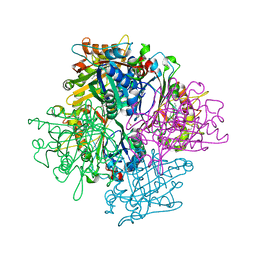 | | DyPB from Rhodococcus jostii RHA1, crystal form 1 | | Descriptor: | DyP Peroxidase, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, R, Roberts, J.N, Grigg, J.C, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1.
Biochemistry, 50, 2011
|
|
3PMB
 
 | |
3P4P
 
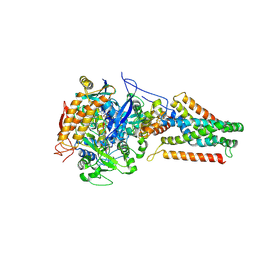 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with fumarate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andr ll, J, Luna-Ch vez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
1A53
 
 | | COMPLEX OF INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS WITH INDOLE-3-GLYCEROLPHOSPHATE AT 2.0 A RESOLUTION | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE, INDOLE-3-GLYCEROLPHOSPHATE SYNTHASE | | Authors: | Hennig, M, Darimont, B, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1998-02-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product.
J.Mol.Biol., 319, 2002
|
|
1AMA
 
 | |
8XG2
 
 | | The structure of HLA-A/Pep14 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XES
 
 | | The structure of HLA-A/L1-1 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
 | | The structure of HLA-A/L1-2 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKE
 
 | | The structure of HLA-A/14-3-D | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | Authors: | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
4XW3
 
 | |
4Y76
 
 | | Factor Xa complex with GTC000401 | | Descriptor: | CALCIUM ION, Coagulation factor X, N~2~-[(6-chloronaphthalen-2-yl)sulfonyl]-N~2~-{(3S)-1-[(2S)-1-(4-methyl-1,4-diazepan-1-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}glycinamide | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
4Y79
 
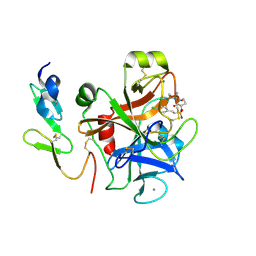 | | Factor Xa complex with GTC000406 | | Descriptor: | (E)-2-(4-chlorophenyl)-N-{(3S)-1-[(2S)-1-(morpholin-4-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}ethenesulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
4YQF
 
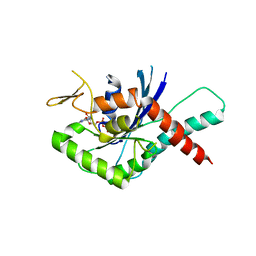 | | GTPase domain of Human Septin 9 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Septin-9 | | Authors: | Matos, S.O, Leonardo, D.A, Macedo, J.N, Pereira, H.M, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | GTPase domain of Human Septin 9
To Be Published
|
|
4YE7
 
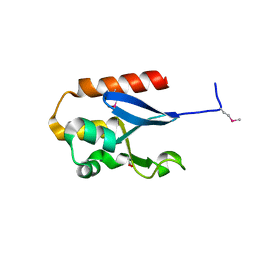 | |
5AF6
 
 | | Structure of Lys33-linked diUb bound to Trabid NZF1 | | Descriptor: | TRABID, UBIQUITIN, ZINC ION | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
5A3K
 
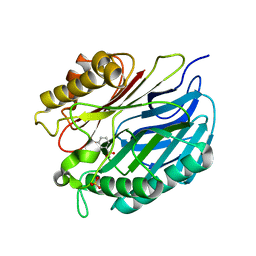 | | Chorismatase mechanisms reveal fundamentally different types of reaction in a single conserved protein fold | | Descriptor: | 3-HYDROXYBENZOIC ACID, PUTATIVE PTERIDINE-DEPENDENT DIOXYGENASE, SULFATE ION | | Authors: | Hubrich, F, Juneja, P, Mueller, M, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
J.Am.Chem.Soc., 137, 2015
|
|
5AF5
 
 | | Structure of Lys33-linked triUb S.G. P 212121 | | Descriptor: | POLYUBIQUITIN-C | | Authors: | Michel, M.A, Elliott, P.R, Swatek, K.N, Simicek, M, Pruneda, J.N, Wagstaff, J.L, Freund, S.M.V, Komander, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Assembly and Specific Recognition of K29- and K33-Linked Polyubiquitin.
Mol.Cell, 58, 2015
|
|
5A97
 
 | | Hazara virus nucleocapsid protain | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Surtees, R, Ariza, A, Hewson, R, Barr, J.N, Edwards, T.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of the Hazara Virus Nucleocapsid Protein.
Bmc Struct.Biol., 15, 2015
|
|
4OX6
 
 | |
4OX8
 
 | |
4N4O
 
 | | Nitrosomonas europea HAO soaked in NH2OH | | Descriptor: | DODECAETHYLENE GLYCOL, HEME C, HYDROXYAMINE, ... | | Authors: | Maalcke, W.J, Dietl, A, Marritt, S.J, Butt, J.N, Jetten, M.S.M, Keltjens, J.T, Barends, T.R.M.B, Kartal, B. | | Deposit date: | 2013-10-08 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structural Basis of Biological NO Generation by Octaheme Oxidoreductases.
J.Biol.Chem., 289, 2014
|
|
4N4M
 
 | | Kuenenia stuttgartiensis hydroxylamine oxidoreductase soaked in phenyl hydrazine | | Descriptor: | 1,2-ETHANEDIOL, 1-PHENYLHYDRAZINE, HEME C, ... | | Authors: | Maalcke, W.J, Dietl, A, Marritt, S.J, Butt, J.N, Jetten, M.S.M, Keltjens, J.T, Barends, T.R.M.B, Kartal, B. | | Deposit date: | 2013-10-08 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Biological NO Generation by Octaheme Oxidoreductases.
J.Biol.Chem., 289, 2014
|
|
4PKI
 
 | |
