2AVW
 
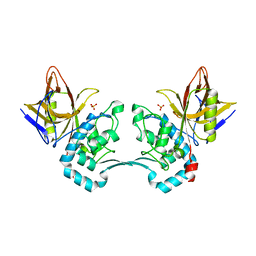 | | Crystal structure of monoclinic form of streptococcus Mac-1 | | Descriptor: | GLYCEROL, IgG-degrading protease, SULFATE ION | | Authors: | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | Deposit date: | 2005-08-30 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
2AX0
 
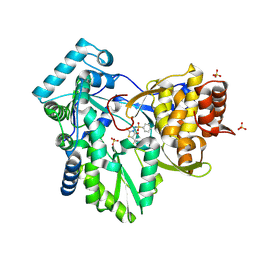 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5x) | | Descriptor: | 5R-(2E-METHYL-3-PHENYL-ALLYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
3CNB
 
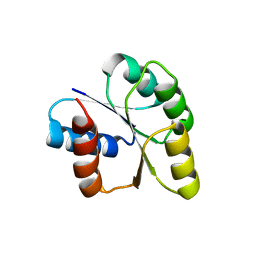 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
2AKM
 
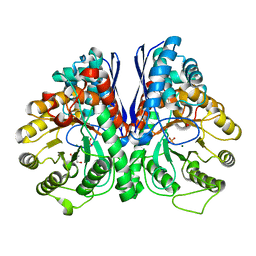 | | Fluoride Inhibition of Enolase: Crystal Structure of the Inhibitory Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gamma enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J.M, Lovelace, L.L. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fluoride inhibition of enolase: crystal structure and thermodynamics
Biochemistry, 45, 2006
|
|
2AWZ
 
 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5h) | | Descriptor: | 5R-(4-BROMOPHENYLMETHYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
2B5G
 
 | | Wild Type SSAT- 1.7A structure | | Descriptor: | Diamine acetyltransferase 1, SULFATE ION | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3D2Y
 
 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3CF5
 
 | | Thiopeptide antibiotic Thiostrepton bound to the large ribosomal subunit of Deinococcus radiodurans | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, 50S RIBOSOMAL PROTEIN L14, ... | | Authors: | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | Deposit date: | 2008-03-02 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Translational Regulation Via L11: Molecular Switches on the Ribosome Turned on and Off by Thiostrepton and Micrococcin.
Mol.Cell, 30, 2008
|
|
2L90
 
 | | Solution structure of murine myristoylated msrA | | Descriptor: | MYRISTIC ACID, Peptide methionine sulfoxide reductase | | Authors: | Gruschus, J.M, Lim, J, Piszczek, G, Levine, R.L, Tjandra, N. | | Deposit date: | 2011-01-27 | | Release date: | 2012-01-11 | | Last modified: | 2012-08-01 | | Method: | SOLUTION NMR | | Cite: | Characterization and solution structure of mouse myristoylated methionine sulfoxide reductase A.
J.Biol.Chem., 287, 2012
|
|
1EQZ
 
 | | X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION | | Descriptor: | 146 NUCLEOTIDES LONG DNA, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Hanson, B.L, Harp, J.M, Timm, D.E, Bunick, G.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Asymmetries in the nucleosome core particle at 2.5 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3VVB
 
 | | Crystal Structure of Capsular Polysaccharide Synthesizing Enzyme CapE from Staphylococcus aureus in apo form | | Descriptor: | CapE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Miyafusa, T, Caaveiro, J.M, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2012-07-18 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the capsular polysaccharide synthesizing protein CapE of Staphylococcus aureus.
Biosci.Rep., 33, 2013
|
|
3V90
 
 | | Structure of T82M glycogenin mutant truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Carrizo, M.E, Romero, J.M, Issoglio, F.M, Curtino, J.A. | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical insight into glycogenin inactivation by the glycogenosis-causing T82M mutation.
Febs Lett., 586, 2012
|
|
3V8Y
 
 | | Structure of apo-glycogenin truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Carrizo, M.E, Romero, J.M, Issoglio, F.M, Curtino, J.A. | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical insight into glycogenin inactivation by the glycogenosis-causing T82M mutation.
Febs Lett., 586, 2012
|
|
3V91
 
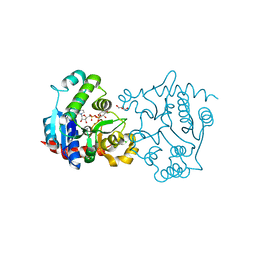 | | Structure of T82M glycogenin mutant truncated at residue 270 complexed with UDP-glucose | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1, ... | | Authors: | Carrizo, M.E, Romero, J.M, Issoglio, F.M, Curtino, J.A. | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical insight into glycogenin inactivation by the glycogenosis-causing T82M mutation.
Febs Lett., 586, 2012
|
|
3VVC
 
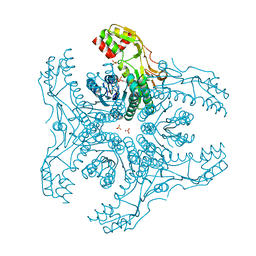 | | Crystal Structure of Capsular Polysaccharide Synthesizing Enzyme CapE , K126E, in apo form | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap8E, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Miyafusa, T, Caaveiro, J.M, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2012-07-18 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the capsular polysaccharide synthesizing protein CapE of Staphylococcus aureus.
Biosci.Rep., 33, 2013
|
|
3W1V
 
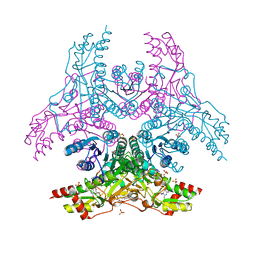 | | Crystal Structure of Capsular Polysaccharide Synthesizing Enzyme CapE from Staphylococcus aureus in complex with inihibitor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Capsular polysaccharide synthesis enzyme Cap8E, SODIUM ION, ... | | Authors: | Miyafusa, T, Caaveiro, J.M, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2012-11-21 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the capsular polysaccharide synthesizing protein CapE of Staphylococcus aureus.
Biosci.Rep., 33, 2013
|
|
1RZ6
 
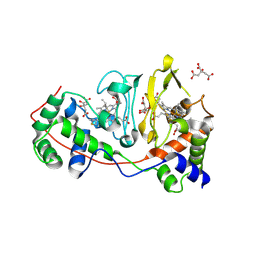 | | Di-haem Cytochrome c Peroxidase, Form IN | | Descriptor: | CITRIC ACID, Cytochrome c peroxidase, HEME C | | Authors: | Dias, J.M, Alves, T, Bonifacio, C, Pereira, A, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617.
Structure, 12, 2004
|
|
3W36
 
 | | Crystal structure of holo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1, VANADATE ION | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
3W35
 
 | | Crystal structure of apo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1 | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
1RZ5
 
 | | Di-haem Cytochrome c Peroxidase, Form OUT | | Descriptor: | CALCIUM ION, Cytochrome c peroxidase, HEME C | | Authors: | Dias, J.M, Alves, T, Bonifacio, C, Pereira, A.S, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617.
Structure, 12, 2004
|
|
3W39
 
 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | Authors: | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | Deposit date: | 2012-12-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
5QD2
 
 | | Crystal structure of BACE complex with BMC017 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(methoxymethyl)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDC
 
 | | Crystal structure of BACE complex with BMC019 hydrolyzed | | Descriptor: | (4S)-4-[(1R)-1,2-dihydroxyethyl]-N,N-dimethyl-2-oxo-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaene-19-carboxamide, Beta-secretase 1, GLYCEROL | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDA
 
 | | Crystal structure of BACE complex with BMC013 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-18-methoxy-3,15,17-triazatricyclo[14.3.1.1~6,10~]henicosa-1(20),6(21),7,9,16,18-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCO
 
 | | Crystal structure of BACE complex with BMC016 | | Descriptor: | (4S)-19-acetyl-4-[(1R)-1-hydroxy-2-({1-[3-(propan-2-yl)phenyl]cyclopropyl}amino)ethyl]-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
