3NO0
 
 | |
3NTB
 
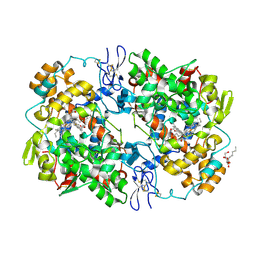 | | Structure of 6-methylthio naproxen analog bound to mCOX-2. | | Descriptor: | (2S)-2-[6-(methylsulfanyl)naphthalen-2-yl]propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3NT1
 
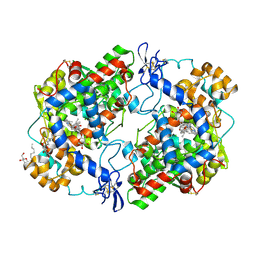 | | High resolution structure of naproxen:COX-2 complex. | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
3NZU
 
 | |
3NZS
 
 | |
3NRZ
 
 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with Hypoxanthine | | Descriptor: | DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cao, H, Pauff, J.M, Hille, R. | | Deposit date: | 2010-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate orientation and catalytic specificity in the action of xanthine oxidase: the sequential hydroxylation of hypoxanthine to uric acid.
J.Biol.Chem., 285, 2010
|
|
3OE3
 
 | | Crystal structure of PliC-St, periplasmic lysozyme inhibitor of C-type lysozyme from Salmonella typhimurium | | Descriptor: | Putative periplasmic protein, SODIUM ION | | Authors: | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2010-08-12 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
3NUH
 
 | |
3O74
 
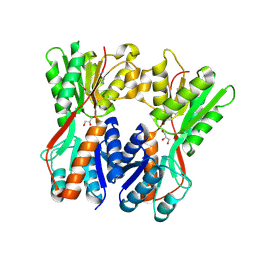 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida | | Descriptor: | Fructose transport system repressor FruR, GLYCEROL | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3NRJ
 
 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a complexed with magnesium | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Had Family Hydrolase from Pseudomonas Syringae Pv.Phaseolica 1448A
To be Published
|
|
3NS1
 
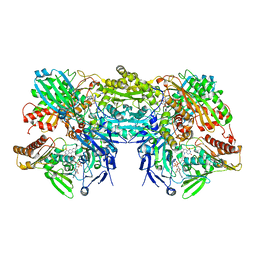 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with 6-Mercaptopurine | | Descriptor: | 9H-purine-6-thiol, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Cao, H, Pauff, J.M, Hille, R. | | Deposit date: | 2010-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate orientation and catalytic specificity in the action of xanthine oxidase: the sequential hydroxylation of hypoxanthine to uric acid.
J.Biol.Chem., 285, 2010
|
|
3OPF
 
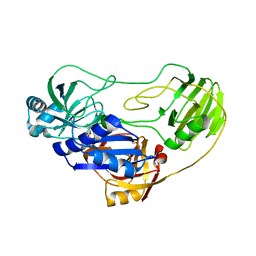 | | Crystal structure of TTHA0988 in space group P212121 | | Descriptor: | GLYCEROL, Putative uncharacterized protein TTHA0988, SULFATE ION | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-01 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OVU
 
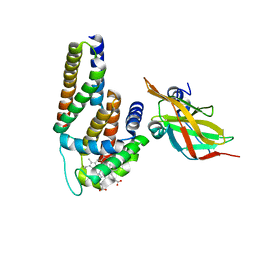 | | Crystal Structure of Human Alpha-Haemoglobin Complexed with AHSP and the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Alpha-hemoglobin-stabilizing protein, Hemoglobin subunit alpha, Iron-regulated surface determinant protein H, ... | | Authors: | Jacques, D.A, Krishna Kumar, K, Caradoc-Davies, T.T, Langley, D.B, Mackay, J.P, Guss, J.M, Gell, D.A. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A new haem pocket structure in alpha-haemoglobin
To be Published
|
|
3OPP
 
 | | ESBL R164S mutant of SHV-1 beta-lactamase complexed with SA2-13 | | Descriptor: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Sampson, J.M, van den Akker, F. | | Deposit date: | 2010-09-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand-dependent disorder of the Omega loop observed in extended-spectrum SHV-type beta-lactamase.
Antimicrob.Agents Chemother., 55, 2011
|
|
3OJY
 
 | | Crystal Structure of Human Complement Component C8 | | Descriptor: | CALCIUM ION, Complement component C8 alpha chain, Complement component C8 beta chain, ... | | Authors: | Lovelace, L.L, Cooper, C.L, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2010-08-23 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement.
J.Biol.Chem., 286, 2011
|
|
3P6D
 
 | |
3OLJ
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) | | Descriptor: | Ribonucleoside-diphosphate reductase subunit M2, SODIUM ION | | Authors: | Chen, X.H, Xu, Z.J, Chen, B.E, Jiang, H.J, Yang, C.G, Zhu, W.L, Shao, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | hRRM2
To be Published
|
|
3OPL
 
 | | ESBL R164H mutant SHV-1 beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Sampson, J.M, van den Akker, F. | | Deposit date: | 2010-09-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand-dependent disorder of the Omega loop observed in extended-spectrum SHV-type beta-lactamase.
Antimicrob.Agents Chemother., 55, 2011
|
|
3OPR
 
 | | ESBL R164H mutant of SHV-1 beta-lactamase complexed to SA2-13 | | Descriptor: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Sampson, J.M, van den Akker, F. | | Deposit date: | 2010-09-01 | | Release date: | 2011-07-13 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ligand-dependent disorder of the Omega loop observed in extended-spectrum SHV-type beta-lactamase.
Antimicrob.Agents Chemother., 55, 2011
|
|
3OQB
 
 | | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110 | | Descriptor: | Oxidoreductase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-02 | | Release date: | 2010-09-15 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110
To be Published
|
|
3P6F
 
 | |
3P6C
 
 | |
3OWO
 
 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
3OTK
 
 | | Structure and mechanisim of core 2 beta1,6-n-acetylglucosaminyltransferase: a Metal-ion independent gt-a glycosyltransferase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase, ... | | Authors: | Pak, J.E, Rini, J.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic characterization of leukocyte-type core 2 beta 1,6-N-acetylglucosaminyltransferase: a metal-ion-independent GT-A glycosyltransferase.
J.Mol.Biol., 414, 2011
|
|
3M2M
 
 | |
