3GG7
 
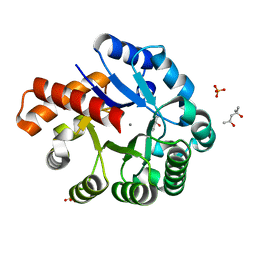 | | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, ... | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans
To be Published
|
|
1O71
 
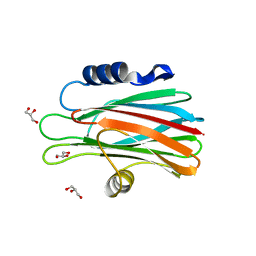 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II complexed with glycerol | | Descriptor: | GLYCEROL, STICHOLYSIN II | | Authors: | Mancheno, J.M, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-10-23 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
1OBT
 
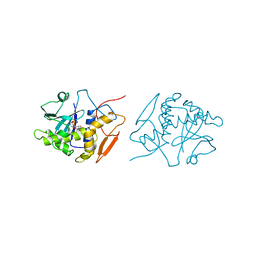 | | STRUCTURE OF RICIN A CHAIN MUTANT, COMPLEX WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-22 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
1OBS
 
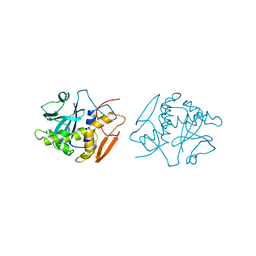 | | STRUCTURE OF RICIN A CHAIN MUTANT | | Descriptor: | RICIN A CHAIN | | Authors: | Day, P.J, Ernst, S.R, Frankel, A.E, Monzingo, A.F, Pascal, J.M, Svinth, M, Robertus, J.D. | | Deposit date: | 1996-06-25 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of an active site substitution of ricin A chain.
Biochemistry, 35, 1996
|
|
1ODN
 
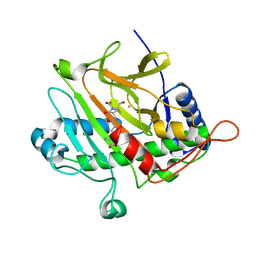 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (OXYGEN-EXPOSED PRODUCT FROM ANAEROBIC AC-VINYLGLYCINE FE COMPLEX) | | Descriptor: | 6-(5-AMINO-5-CARBOXY-PENTANOYLAMINO)-3-HYDROXYMETHYL-7-OXO-4-THIA-1-AZA-BICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Elkins, J.M, Rutledge, P.J, Burzlaff, N.I, Clifton, I.J, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Studies on the Reaction of Isopenicillin N Synthase with an Unsaturated Substrate Analogue
Org.Biomol.Chem., 1, 2003
|
|
3DXS
 
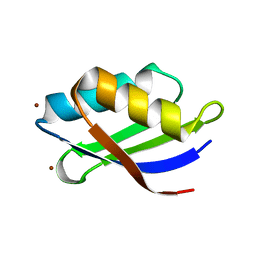 | | Crystal structure of a copper binding domain from HMA7, a P-type ATPase | | Descriptor: | Copper-transporting ATPase RAN1, LITHIUM ION, ZINC ION | | Authors: | Zimmermann, M, Xiao, Z, Clarke, O.B, Gulbis, J.M, Wedd, A.G. | | Deposit date: | 2008-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains.
Biochemistry, 48, 2009
|
|
1ODM
 
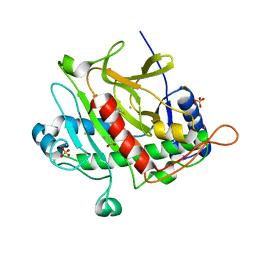 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ANAEROBIC AC-VINYLGLYCINE FE COMPLEX) | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-VINYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Elkins, J.M, Rutledge, P.J, Burzlaff, N.I, Clifton, I.J, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic Studies on the Reaction of Isopenicillin N Synthase with an Unsaturated Substrate Analogue
Org.Biomol.Chem., 1, 2003
|
|
1OW6
 
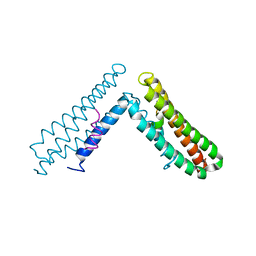 | | Paxillin LD4 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Campbell, I.D, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Recognition of Paxillin LD motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
1P1T
 
 | |
3DXR
 
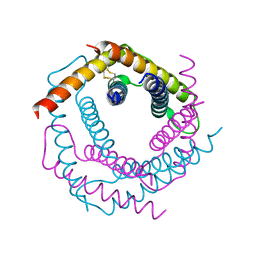 | |
1OMZ
 
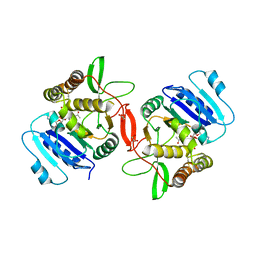 | | crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2),
a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1P7B
 
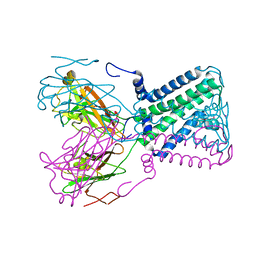 | | Crystal structure of an inward rectifier potassium channel | | Descriptor: | POTASSIUM ION, integral membrane channel and cytosolic domains | | Authors: | Kuo, A, Gulbis, J.M, Antcliff, J.F, Rahman, T, Lowe, E.D, Zimmer, J, Cuthbertson, J, Ashcroft, F.M, Ezaki, T, Doyle, D.A. | | Deposit date: | 2003-05-01 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structure of the potassium channel KirBac1.1 in the closed state.
Science, 300, 2003
|
|
1OSV
 
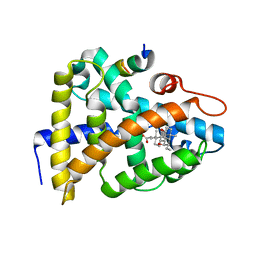 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1OMX
 
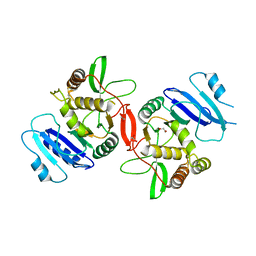 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2 | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1P33
 
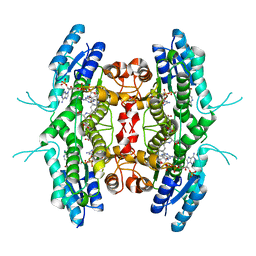 | | Pteridine reductase from Leishmania tarentolae complex with NADPH and MTX | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Zhao, H, Bray, T, Ouellette, M, Zhao, M, Ferre, R.A, Matthews, D, Whiteley, J.M, Varughese, K.I. | | Deposit date: | 2003-04-16 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of pteridine reductase (PTR1) from Leishmania tarentolae.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3GB5
 
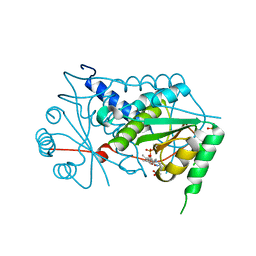 | | Crystal structure of Mus musculus iodotyrosine deiodinase (IYD) bound to FMN | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Iodotyrosine dehalogenase 1, ... | | Authors: | Thomas, S.R, McTamney, P.M, Adler, J.M, LaRonde-LeBlanc, N, Rokita, S.E. | | Deposit date: | 2009-02-18 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands.
J.Biol.Chem., 284, 2009
|
|
1P44
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | 5-{[4-(9H-FLUOREN-9-YL)PIPERAZIN-1-YL]CARBONYL}-1H-INDOLE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
1P4Y
 
 | |
1P9P
 
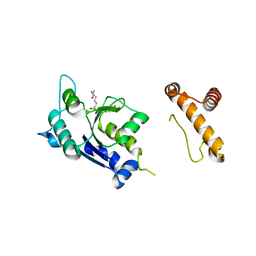 | | The Crystal Structure of a M1G37 tRNA Methyltransferase, TrmD | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Watts, J.M, Zalacain, M, Van Thiel, A, Vitaszka, P.R, Redlak, M, Andraos-Selim, C, Rastinejad, F, Holmes, W.M. | | Deposit date: | 2003-05-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Catalysis by a Knotted TrmD tRNA Methyltransferase.
J.Mol.Biol., 333, 2003
|
|
3GHF
 
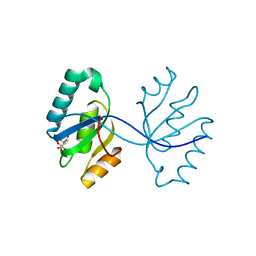 | | Crystal structure of the septum site-determining protein minC from Salmonella typhimurium | | Descriptor: | CITRIC ACID, Septum site-determining protein minC | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the septum site-determining protein minC from Salmonella typhimurium
To be Published
|
|
1PVO
 
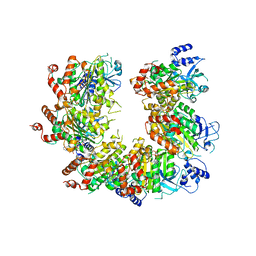 | |
3G8R
 
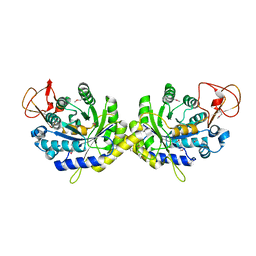 | | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472 | | Descriptor: | Probable spore coat polysaccharide biosynthesis protein E, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-12 | | Release date: | 2009-04-07 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472
To be Published
|
|
3G5L
 
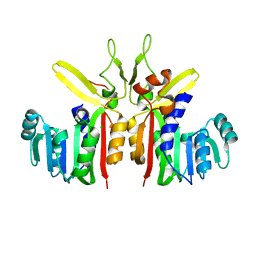 | | Crystal structure of putative S-adenosylmethionine dependent methyltransferase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Putative S-adenosylmethionine dependent methyltransferase | | Authors: | Patskovsky, Y, Sampathkumar, P, Gilmore, M, Miller, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of S-Adenosylmethionine Dependent Methyltransferase from Listeria Monocytogenes
To be Published
|
|
3FYR
 
 | | Crystal structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis | | Descriptor: | Sporulation inhibitor sda | | Authors: | Jacques, D.A, Streamer, M, King, G.F, Guss, J.M, Trewhella, J, Langley, D.B. | | Deposit date: | 2009-01-23 | | Release date: | 2009-06-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3G1T
 
 | | CRYSTAL STRUCTURE OF short chain dehydrogenase from Salmonella enterica subsp. enterica serovar Typhi str. CT18 | | Descriptor: | MAGNESIUM ION, short chain dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF short chain dehydrogenase from Salmonella enterica subsp. enterica
serovar Typhi str. CT18
To be Published
|
|
