1DL8
 
 | | CRYSTAL STRUCTURE OF 5-F-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | 分子名称: | 5-FLUORO-9-AMINO-(N-(2-DIMETHYLAMINO)ETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | 著者 | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P. | | 登録日 | 1999-12-08 | | 公開日 | 2000-10-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Acridinecarboxamide topoisomerase poisons: structural and kinetic studies of the DNA complexes of 5-substituted 9-amino-(N-(2-dimethylamino)ethyl)acridine-4-carboxamides.
Mol.Pharmacol., 58, 2000
|
|
7TJF
 
 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA, 84 bp ARS1, ... | | 著者 | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | 登録日 | 2022-01-16 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
1DG2
 
 | | SOLUTION CONFORMATION OF A-CONOTOXIN AUIB | | 分子名称: | A-CONOTOXIN AUIB | | 著者 | Cho, J.-H, Mok, K.H, Olivera, B.M, McIntosh, J.M, Park, K.-H, Han, K.-H. | | 登録日 | 1999-11-23 | | 公開日 | 2000-02-25 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nuclear magnetic resonance solution conformation of alpha-conotoxin AuIB, an alpha(3)beta(4) subtype-selective neuronal nicotinic acetylcholine receptor antagonist.
J.Biol.Chem., 275, 2000
|
|
1DD6
 
 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | 分子名称: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | 著者 | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | 登録日 | 1999-11-08 | | 公開日 | 2000-11-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DB1
 
 | | CRYSTAL STRUCTURE OF THE NUCLEAR RECEPTOR FOR VITAMIN D COMPLEXED TO VITAMIN D | | 分子名称: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D NUCLEAR RECEPTOR | | 著者 | Rochel, N, Wurtz, J.M, Mitschler, A, Klaholz, B, Moras, D. | | 登録日 | 1999-11-02 | | 公開日 | 2000-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand.
Mol.Cell, 5, 2000
|
|
1DD9
 
 | | STRUCTURE OF THE DNAG CATALYTIC CORE | | 分子名称: | DNA PRIMASE, STRONTIUM ION | | 著者 | Keck, J.L, Roche, D.D, Lynch, A.S, Berger, J.M. | | 登録日 | 1999-11-09 | | 公開日 | 2000-04-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of the RNA polymerase domain of E. coli primase.
Science, 287, 2000
|
|
1DGP
 
 | | ARISTOLOCHENE SYNTHASE FARNESOL COMPLEX | | 分子名称: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, ARISTOLOCHENE SYNTHASE | | 著者 | Caruthers, J.M, Kang, I, Cane, D.E, Christianson, D.W. | | 登録日 | 1999-11-24 | | 公開日 | 2001-02-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti.
J.Biol.Chem., 275, 2000
|
|
5QD2
 
 | | Crystal structure of BACE complex with BMC017 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-19-(methoxymethyl)-11,16-dioxa-3-azatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1DCW
 
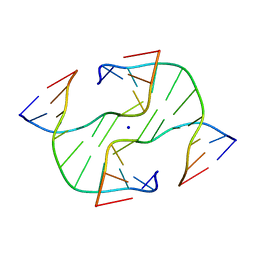 | | STRUCTURE OF A FOUR-WAY JUNCTION IN AN INVERTED REPEAT SEQUENCE. | | 分子名称: | DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3'), SODIUM ION | | 著者 | Eichman, B.F, Vargason, J.M, Mooers, B.H.M, Ho, P.S. | | 登録日 | 1999-11-05 | | 公開日 | 2000-04-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Holliday junction in an inverted repeat DNA sequence: sequence effects on the structure of four-way junctions.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DDB
 
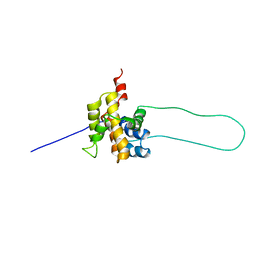 | | STRUCTURE OF MOUSE BID, NMR, 20 STRUCTURES | | 分子名称: | PROTEIN (BID) | | 著者 | Mcdonnell, J.M, Fushman, D, Milliman, C, Korsmeyer, S.J, Cowburn, D. | | 登録日 | 1999-02-19 | | 公開日 | 1999-08-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists.
Cell(Cambridge,Mass.), 96, 1999
|
|
5QDA
 
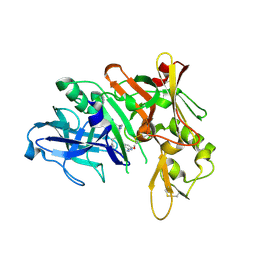 | | Crystal structure of BACE complex with BMC013 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-18-methoxy-3,15,17-triazatricyclo[14.3.1.1~6,10~]henicosa-1(20),6(21),7,9,16,18-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QCO
 
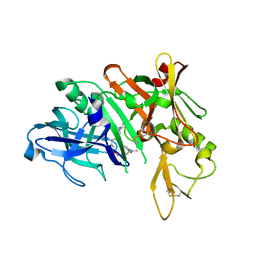 | | Crystal structure of BACE complex with BMC016 | | 分子名称: | (4S)-19-acetyl-4-[(1R)-1-hydroxy-2-({1-[3-(propan-2-yl)phenyl]cyclopropyl}amino)ethyl]-11-oxa-3,16-diazatricyclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
7UDZ
 
 | | Designed pentameric proton channel LQLL | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel LQLL | | 著者 | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J.M, Liu, L, DeGrado, W.F. | | 登録日 | 2022-03-20 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
5QDD
 
 | | Crystal structure of BACE complex with BMC020 hydrolyzed | | 分子名称: | (10R,12S)-12-[(1R)-1,2-dihydroxyethyl]-N,N,10-trimethyl-14-oxo-2-oxa-13-azabicyclo[13.3.1]nonadeca-1(19),15,17-triene-17-carboxamide, Beta-secretase 1, GLYCEROL | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1DI2
 
 | |
5QD3
 
 | | Crystal structure of BACE complex with BMC010 | | 分子名称: | (10R,12S)-12-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-17-(methoxymethyl)-10-methyl-2,13-diazabicyclo[13.3.1]nonadeca-1(19),15,17-trien-14-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
1DKI
 
 | | CRYSTAL STRUCTURE OF THE ZYMOGEN FORM OF STREPTOCOCCAL PYROGENIC EXOTOXIN B ACTIVE SITE (C47S) MUTANT | | 分子名称: | PYROGENIC EXOTOXIN B ZYMOGEN, SULFATE ION | | 著者 | Kagawa, T.F, Cooney, J.C, Baker, H.M, McSweeney, S, Liu, M, Gubba, S, Musser, J.M, Baker, E.N. | | 登録日 | 1999-12-07 | | 公開日 | 2000-03-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: an integrin-binding cysteine protease.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DML
 
 | | CRYSTAL STRUCTURE OF HERPES SIMPLEX UL42 BOUND TO THE C-TERMINUS OF HSV POL | | 分子名称: | DNA POLYMERASE, DNA POLYMERASE PROCESSIVITY FACTOR | | 著者 | Zuccola, H.J, Filman, D.J, Coen, D.M, Hogle, J.M. | | 登録日 | 1999-12-14 | | 公開日 | 2000-03-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase.
Mol.Cell, 5, 2000
|
|
1DSX
 
 | | KV1.2 T1 DOMAIN, RESIDUES 33-119, T46V MUTANT | | 分子名称: | PROTEIN (KV1.2 VOLTAGE-GATED POTASSIUM CHANNEL) | | 著者 | Minor Jr, D.L, Lin, Y.-F, Mobley, B.C, Avelar, A, Jan, Y.N, Jan, L.Y, Berger, J.M. | | 登録日 | 2000-01-10 | | 公開日 | 2000-09-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The polar T1 interface is linked to conformational changes that open the voltage-gated potassium channel.
Cell(Cambridge,Mass.), 102, 2000
|
|
1DL5
 
 | | PROTEIN-L-ISOASPARTATE O-METHYLTRANSFERASE | | 分子名称: | CADMIUM ION, CHLORIDE ION, PROTEIN-L-ISOASPARTATE O-METHYLTRANSFERASE, ... | | 著者 | Skinner, M.M, Puvathingal, J.M, Walter, R.L, Friedman, A.M. | | 登録日 | 1999-12-08 | | 公開日 | 2000-12-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of protein isoaspartyl methyltransferase: a catalyst for protein repair.
Structure Fold.Des., 8, 2000
|
|
3IZG
 
 | | Bacteriophage T7 prohead shell EM-derived atomic model | | 分子名称: | Major capsid protein 10A | | 著者 | Ionel, A, Velazquez-Muriel, J.A, Agirrezabala, X, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | 登録日 | 2010-10-27 | | 公開日 | 2010-11-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (10.9 Å) | | 主引用文献 | Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
7UCR
 
 | | Joint X-ray/neutron structure of the Sarcin-Ricin loop RNA | | 分子名称: | SULFATE ION, Sarcin-Ricin loop RNA | | 著者 | Harp, J.M, Egli, M.E, Pallan, P.S, Coates, L. | | 登録日 | 2022-03-17 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1 Å), X-RAY DIFFRACTION | | 主引用文献 | Cryo neutron crystallography demonstrates influence of RNA 2'-OH orientation on conformation, sugar pucker and water structure.
Nucleic Acids Res., 50, 2022
|
|
7UKS
 
 | | Crystal structure of SOS1 with phthalazine inhibitor bound (compound 15) | | 分子名称: | 4-methyl-N-{(1R)-1-[2-methyl-3-(trifluoromethyl)phenyl]ethyl}-7-(piperazin-1-yl)phthalazin-1-amine, Son of sevenless homolog 1 | | 著者 | Gunn, R.J, Lawson, J.D, Ketcham, J.M, Marx, M.A. | | 登録日 | 2022-04-01 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Design and Discovery of MRTX0902, a Potent, Selective, Brain-Penetrant, and Orally Bioavailable Inhibitor of the SOS1:KRAS Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
7UKR
 
 | | Crystal Structure of SOS1 with MRTX0902, a Potent and Selective Inhibitor of the SOS1:KRAS Protein-Protein Interaction | | 分子名称: | 2-methyl-3-[(1R)-1-{[4-methyl-7-(morpholin-4-yl)pyrido[3,4-d]pyridazin-1-yl]amino}ethyl]benzonitrile, Son of sevenless homolog 1 | | 著者 | Gunn, R.J, Lawson, J.D, Ketcham, J.M, Marx, M.A. | | 登録日 | 2022-04-01 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design and Discovery of MRTX0902, a Potent, Selective, Brain-Penetrant, and Orally Bioavailable Inhibitor of the SOS1:KRAS Protein-Protein Interaction.
J.Med.Chem., 65, 2022
|
|
5QCY
 
 | | Crystal structure of BACE complex with BMC008 | | 分子名称: | (9R,11S)-11-[(1R)-1-hydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-9-methyl-16-(1,3-oxazol-2-yl)-3-[(1R)-1-phenylethyl]-3,12-diazabicyclo[12.3.1]octadeca-1(18),14,16-triene-2,13-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
