4K0Y
 
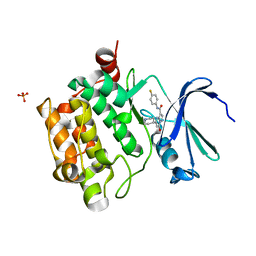 | | Structure of PIM-1 kinase bound to N-(4-fluorophenyl)-7-hydroxy-5-(piperidin-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Descriptor: | N-(4-fluorophenyl)-7-hydroxy-5-(piperidin-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H, Steffek, M. | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Discovery of novel pyrazolo[1,5-a]pyrimidines as potent pan-Pim inhibitors by structure- and property-based drug design.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JUE
 
 | |
8WZ8
 
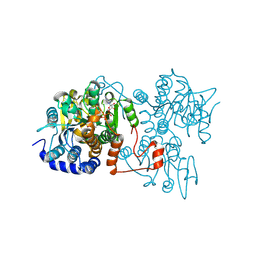 | | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(T67A,Q69A) complex with NAD | | Descriptor: | Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Gao, Y.S, Xie, R, Chen, Y.N, Ma, J.M, Ge, H.H. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | The crystal structure of Legionella pneumophila adenosylhomocysteinase Lpg2021(T67A,Q69A) complex with NAD
To Be Published
|
|
1BIK
 
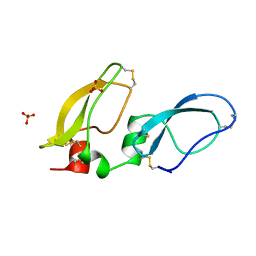 | | X-RAY STRUCTURE OF BIKUNIN FROM THE HUMAN INTER-ALPHA-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIKUNIN, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Guss, J.M, Ollis, D.L. | | Deposit date: | 1997-11-26 | | Release date: | 1999-03-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of bikunin from the inter-alpha-inhibitor complex: a serine protease inhibitor with two Kunitz domains.
J.Mol.Biol., 276, 1998
|
|
1BE7
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN C42S MUTANT | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Maher, M, Guss, J.M, Wilce, M, Wedd, A.G. | | Deposit date: | 1998-05-20 | | Release date: | 1998-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Rubredoxin from Clostridium Pasteurianum: Mutation of the Iron Cysteinyl Ligands to Serine. Crystal and Molecular Structures of the Oxidised and Dithionite-Treated Forms of the Cys42Ser Mutant
J.Am.Chem.Soc., 120, 1998
|
|
4J6I
 
 | | Discovery of thiazolobenzoxepin PI3-kinase inhibitors that spare the PI3-kinase beta isoform | | Descriptor: | 2-methyl-1-(4-{2-[1-(2,2,2-trifluoroethyl)-1H-1,2,4-triazol-5-yl]-4,5-dihydro[1]benzoxepino[5,4-d][1,3]thiazol-8-yl}-1H-pyrazol-1-yl)propan-2-ol, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Rouge, L, Wu, P. | | Deposit date: | 2013-02-11 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of thiazolobenzoxepin PI3-kinase inhibitors that spare the PI3-kinase beta isoform.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1BB8
 
 | |
1BI5
 
 | | CHALCONE SYNTHASE FROM ALFALFA | | Descriptor: | CHALCONE SYNTHASE | | Authors: | Ferrer, J.L, Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1BGW
 
 | | TOPOISOMERASE RESIDUES 410-1202, | | Descriptor: | TOPOISOMERASE | | Authors: | Berger, J.M, Gamblin, S.J, Harrison, S.C, Wang, J.C. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of DNA topoisomerase II.
Nature, 379, 1996
|
|
4EZJ
 
 | |
3T8M
 
 | |
3TDM
 
 | | Computationally designed TIM-barrel protein, HalfFLR | | Descriptor: | Computationally designed two-fold symmetric TIM-barrel protein, FLR (half molecule), PHOSPHATE ION | | Authors: | Harp, J.M, Fortenberry, C, Bowman, E, Profitt, W, Dorr, B, Mizoue, L. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploring symmetry as an avenue to the computational design of large protein domains.
J.Am.Chem.Soc., 133, 2011
|
|
8J4N
 
 | |
8J4O
 
 | |
8VGA
 
 | |
8VGB
 
 | |
1AG7
 
 | | CONOTOXIN GS, NMR, 20 STRUCTURES | | Descriptor: | CONOTOXIN GS | | Authors: | Hill, J.M, Alewood, P.F, Craik, D.J. | | Deposit date: | 1997-04-03 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sodium channel antagonist conotoxin GS: a new molecular caliper for probing sodium channel geometry.
Structure, 5, 1997
|
|
4GMY
 
 | |
1A16
 
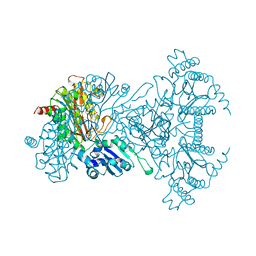 | | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | | Descriptor: | AMINOPEPTIDASE P, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilce, M.C, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
183D
 
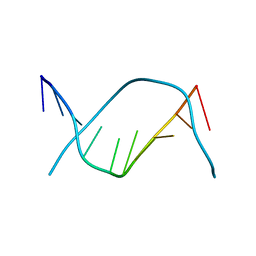 | | X-RAY STRUCTURE OF A DNA DECAMER CONTAINING 7, 8-DIHYDRO-8-OXOGUANINE | | Descriptor: | DNA (5'-D(*CP*CP*AP*(8OG)P*CP*GP*CP*TP*GP*G)-3') | | Authors: | Lipscomb, L.A, Peek, M.E, Morningstar, M.L, Verghis, S.M, Miller, E.M, Rich, A, Essigmann, J.M, Williams, L.D. | | Deposit date: | 1994-08-01 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
4E6Q
 
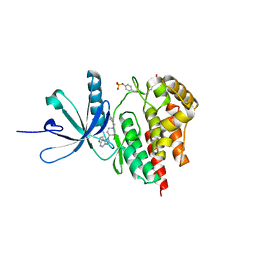 | | JAK2 kinase (JH1 domain) triple mutant in complex with compound 12 | | Descriptor: | 1-(1-benzylpiperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK2 | | Authors: | Murray, J.M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-30 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
1B79
 
 | |
1B2J
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B34
 
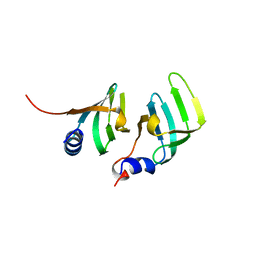 | | CRYSTAL STRUCTURE OF THE D1D2 SUB-COMPLEX FROM THE HUMAN SNRNP CORE DOMAIN | | Descriptor: | PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN SM D1), PROTEIN (SMALL NUCLEAR RIBONUCLEOPROTEIN SM D2) | | Authors: | Walke, S, Young, R.J, Kambach, C, Avis, J.M, De La Fortelle, E, Li, J, Nagai, K. | | Deposit date: | 1998-12-17 | | Release date: | 2000-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs.
Cell(Cambridge,Mass.), 96, 1999
|
|
1BIW
 
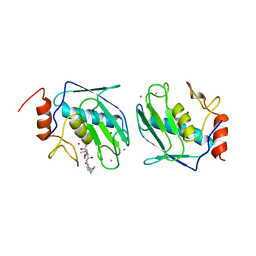 | | DESIGN AND SYNTHESIS OF CONFORMATIONALLY-CONSTRAINED MMP INHIBITORS | | Descriptor: | CALCIUM ION, N1-HYDROXY-2-(3-HYDROXY-PROPYL)-3-ISOBUTYL-N4-[1-(2-METHOXY-ETHYL)-2-OXO-AZEPAN-3-YL]-SUCCINAMIDE, PROTEIN (STROMELYSIN-1 COMPLEX), ... | | Authors: | Natchus, M.G, Cheng, M, Wahl, C.T, Pikul, S, Almstead, N.G, Bradley, R.S, Taiwo, Y.O, Mieling, G.E, Dunaway, C.M, Snider, C.E, McIver, J.M, Barnett, B.L, McPhail, S.J, Anastasio, M.B, De, B. | | Deposit date: | 1998-06-19 | | Release date: | 1999-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and synthesis of conformationally-constrained MMP inhibitors.
Bioorg.Med.Chem.Lett., 8, 1998
|
|
