8PGR
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3270 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(2-methoxypyridin-4-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3270
To Be Published
|
|
8PG7
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2755 | | Descriptor: | 7-[(1~{S})-1-[3-(4-azanylbutyl)-2,5-bis(oxidanylidene)imidazolidin-1-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2755
To Be Published
|
|
8PG6
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2754 | | Descriptor: | 7-[(1~{S})-1-[3-(3-azanylpropyl)-2,5-bis(oxidanylidene)imidazolidin-1-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2754
To Be Published
|
|
8PGI
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2984 | | Descriptor: | 7-[(1~{S})-1-[5-[(3-azanyl-3-methyl-azetidin-1-yl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2984
To Be Published
|
|
8PGY
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3462 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(6-methoxy-5-oxidanyl-pyridin-3-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3462
To Be Published
|
|
8PGO
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3183 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylamino)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3183
To Be Published
|
|
8PG5
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2731 | | Descriptor: | 3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-7-[(1~{S})-1-(piperidin-4-ylmethoxy)ethyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2731
To Be Published
|
|
8PGF
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2941 | | Descriptor: | 7-[(1~{S})-1-[[4-(aminomethyl)phenyl]carbonylamino]ethyl]-3-[6-(morpholin-4-ylmethyl)pyridin-3-yl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2941
To Be Published
|
|
8PGL
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3027 | | Descriptor: | 7-[(1~{S})-1-[5-(carbamimidamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3027
To Be Published
|
|
8PGM
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3062 | | Descriptor: | 3-[4-[(azanyl-methyl-oxidanyl-$l^{4}-sulfanyl)methyl]-3-fluoranyl-phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3062
To Be Published
|
|
8PGZ
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3528 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(6-oxidanylidene-1~{H}-pyridazin-4-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3528
To Be Published
|
|
1XXQ
 
 | | Structure of a mannose-specific jacalin-related lectin from Morus nigra | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Rabijns, A, Barre, A, Van Damme, E.J.M, Peumans, W.J, De Ranter, C.J, Rouge, P. | | Deposit date: | 2004-11-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the jacalin-related lectin MornigaM from the black mulberry (Morus nigra) in complex with mannose
Febs J., 272, 2005
|
|
4N6B
 
 | | Soybean Serine Acetyltransferase Complexed with CoA | | Descriptor: | COENZYME A, Serine Acetyltransferase Apoenzyme | | Authors: | Yi, H, Dey, S, Kumaran, S, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone.
J.Biol.Chem., 288, 2013
|
|
5I7Y
 
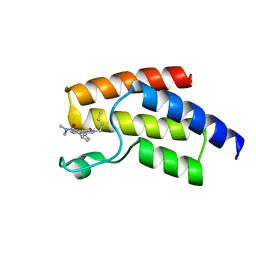 | | BRD9 in complex with Cpd4 ((E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide) | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Bromodomain-containing protein 9 | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4514 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
5I80
 
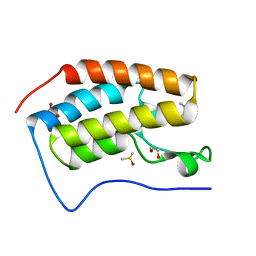 | | BRD4 in complex with Cpd2 (N,N-dimethyl-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)benzamide) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4501 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
4N5C
 
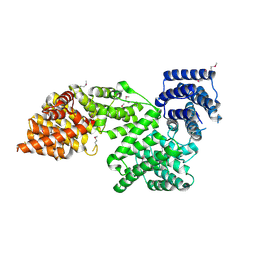 | | Crystal structure of Ypp1 | | Descriptor: | Cargo-transport protein YPP1 | | Authors: | Wu, X, Chi, R.J, Baskin, J.M, Lucast, L, Burd, C.G, De Camilli, P, Reinisch, K.M. | | Deposit date: | 2013-10-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex.
Dev.Cell, 28, 2014
|
|
4N5A
 
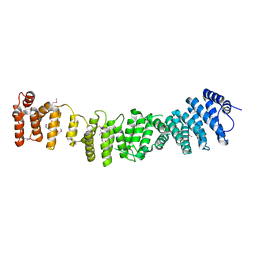 | | Crystal structure of Efr3 | | Descriptor: | Protein EFR3 | | Authors: | Wu, X, Chi, R.J, Baskin, J.M, Lucast, L, Burd, C.G, De Camilli, P, Reinisch, K.M. | | Deposit date: | 2013-10-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex.
Dev.Cell, 28, 2014
|
|
4N49
 
 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with m7GpppG and SAM | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
2HHL
 
 | | Crystal structure of the human small CTD phosphatase 3 isoform 1 | | Descriptor: | 12-TUNGSTOPHOSPHATE, CTD small phosphatase-like protein | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4MX5
 
 | | Structure of ricin A chain bound with benzyl-(2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)ethyl)carbamate | | Descriptor: | Ricin A chain, benzyl (2-{[(2-amino-4-oxo-3,4-dihydropteridin-7-yl)carbonyl]amino}ethyl)carbamate | | Authors: | Jasheway, K.R, Robertus, J.D, Wiget, P.A, Manzano, L.A, Pruet, J.M, Gao, G, Saito, R, Monzingo, A.F, Anslyn, E.V. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Sulfur incorporation generally improves Ricin inhibition in pterin-appended glycine-phenylalanine dipeptide mimics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1MBT
 
 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the substrate-free form of MurB, an essential enzyme for the synthesis of bacterial cell walls.
Structure, 4, 1996
|
|
5H3C
 
 | |
4ON0
 
 | | Crystal Structure of NolR from Sinorhizobium fredii in complex with oligo AA DNA | | Descriptor: | DNA (5 -D(*TP*AP*AP*TP*CP*TP*CP*TP*TP*GP*GP*GP*AP*CP*TP*AP*CP*AP*AP*TP*TP*A)-3 ), DNA (5 -D(*TP*AP*TP*TP*AP*GP*AP*GP*AP*AP*CP*CP*CP*TP*GP*AP*TP*GP*TP*TP*AP*A)-3 ), NolR | | Authors: | Lee, S.G, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for regulation of rhizobial nodulation and symbiosis gene expression by the regulatory protein NolR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P2B
 
 | |
2GSH
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium | | Descriptor: | GLYCEROL, L-RHAMNONATE DEHYDRATASE, MAGNESIUM ION | | Authors: | Patskovsky, Y, Malashkevich, V.N, Sauder, J.M, Dickey, M, Adams, J.M, Wasserman, S.R, Gerlt, J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal Structure of L-rhamnonate dehydratase from Salmonella Typhimurium Lt2
To be Published
|
|
