1OXX
 
 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION | | Authors: | Verdon, G, Albers, S.-V, van Oosterwijk, N, Dijkstra, B.W, Driessen, A.J.M, Thunnissen, A.M.W.H. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus
J.Mol.Biol., 334, 2003
|
|
4HTT
 
 | |
4TSO
 
 | | Crystal structure of FraC with DHPC bound (crystal form I) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Fragaceatoxin C, PHOSPHATE ION, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4KS1
 
 | | Influenza neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-amino-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-amino-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
4TSP
 
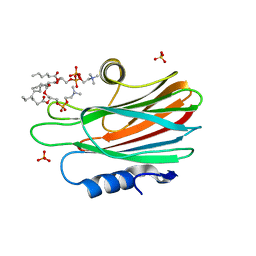 | | Crystal structure of FraC with DHPC bound (crystal form II) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Fragaceatoxin C, PHOSPHATE ION, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSY
 
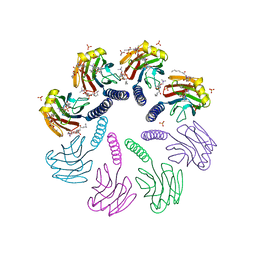 | | Crystal structure of FraC with lipids | | Descriptor: | 2-[[(E,2S,3R)-2-(hexanoylamino)-3-oxidanyl-dec-4-enoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, Fragaceatoxin C, HEPTANE-1,2,3-TRIOL, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSL
 
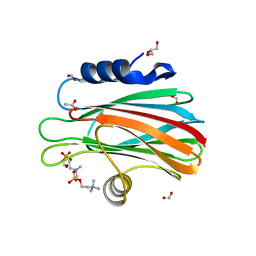 | | Crystal structure of FraC with POC bound (crystal form I) | | Descriptor: | ACETATE ION, FORMIC ACID, Fragaceatoxin C, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TQG
 
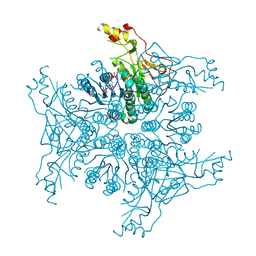 | | Crystal structure of Megavirus UDP-GlcNAc 4,6-dehydratase, 5-epimerase Mg534 | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative dTDP-d-glucose 4 6-dehydratase | | Authors: | Jeudy, S, Piacente, F, De Castro, C, Molinaro, A, Salis, A, Damonte, G, Bernardi, C, Tonetti, M, Claverie, J.M, Abergel, C. | | Deposit date: | 2014-06-11 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Giant Virus Megavirus chilensis Encodes the Biosynthetic Pathway for Uncommon Acetamido Sugars.
J.Biol.Chem., 289, 2014
|
|
4TSN
 
 | | Crystal structure of FraC with POC bound (crystal form II) | | Descriptor: | ACETATE ION, Fragaceatoxin C, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSQ
 
 | | Crystal structure of FraC with DHPC bound (crystal form III) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, Fragaceatoxin C, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
1SCY
 
 | |
1A3K
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN GALECTIN-3 CARBOHYDRATE RECOGNITION DOMAIN (CRD) AT 2.1 ANGSTROM RESOLUTION | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seetharaman, J, Kanigsberg, A, Slaaby, R, Leffler, H, Barondes, S.H, Rini, J.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution.
J.Biol.Chem., 273, 1998
|
|
4KS3
 
 | | Influenza Neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-[4-(3-hydroxypropyl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-[4-(3-hydroxypropyl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
4KS5
 
 | | Influenza neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-[4-(2-hydroxypropan-2-yl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-[4-(2-hydroxypropan-2-yl)-1H-1,2,3-triazol-1-yl]-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
3FZW
 
 | | Crystal Structure of Ketosteroid Isomerase D40N-D103N from Pseudomonas putida (pKSI) with bound equilenin | | Descriptor: | EQUILENIN, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Caaveiro, J.M.M, Ringe, D, Petsko, G.A. | | Deposit date: | 2009-01-26 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Hydrogen bond coupling in the ketosteroid isomerase active site.
Biochemistry, 48, 2009
|
|
1AFS
 
 | | RECOMBINANT RAT LIVER 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE (3-ALPHA-HSD) COMPLEXED WITH NADP AND TESTOSTERONE | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TESTOSTERONE | | Authors: | Bennett, M.J, Albert, R.H, Jez, J.M, Ma, H, Penning, T.M, Lewis, M. | | Deposit date: | 1997-03-13 | | Release date: | 1997-10-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
Structure, 5, 1997
|
|
3HKX
 
 | |
4UC5
 
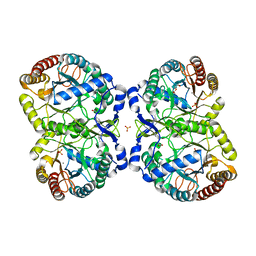 | | Neisseria Meningitidis DAH7PS-Phenylalanine regulated | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Heyes, L.C, Lang, E.J.M, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Calculated Pka Variations Expose Dynamic Allosteric Communication Networks.
J.Am.Chem.Soc., 138, 2016
|
|
3F8F
 
 | | Crystal structure of multidrug binding transcriptional regulator LmrR complexed with Daunomycin | | Descriptor: | DAUNOMYCIN, Transcriptional regulator, PadR-like family | | Authors: | Madoori, P.K, Agustiandari, H, Driessen, A.J.M, Thunnissen, A.-M.W.H. | | Deposit date: | 2008-11-12 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition.
Embo J., 28, 2009
|
|
1T65
 
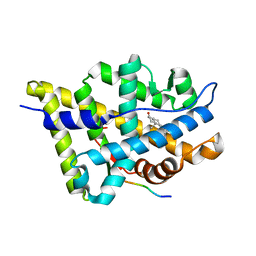 | | Crystal structure of the androgen receptor ligand binding domain with DHT and a peptide derived form its physiological coactivator GRIP1 NR box 2 bound in a non-helical conformation | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 2 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
4KY1
 
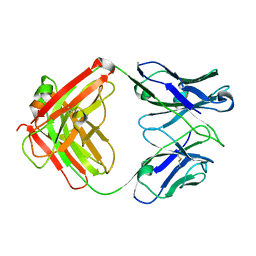 | | humanized HP1/2 Fab | | Descriptor: | IMMUNOGLOBULIN IGG1 FAB, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Arndt, J.W, Hanf, K.J.M, Lugovskoy, A, Chen, L.L, Jarpe, M, Boriack-Sjodin, A, Li, Y, van Vlijmen, H, Pepinsky, B, Taylor, F, Silvian, L, Taveras, A. | | Deposit date: | 2013-05-28 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Antibody humanization by redesign of complementarity-determining region residues proximate to the acceptor framework.
METHODS (SAN DIEGO), 65, 2014
|
|
1BPS
 
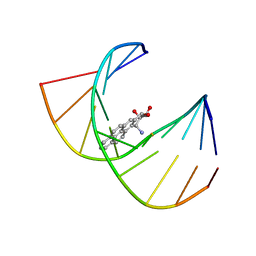 | | MINOR CONFORMER OF A BENZO[A]PYRENE DIOL EPOXIDE ADDUCT OF DA IN DUPLEX DNA | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA (5'-D(*CP*TP*CP*GP*GP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*(BAP)AP*CP*GP*AP*G)-3') | | Authors: | Schwartz, J.S, Rice, J.S, Luxon, B.A, Sayer, J.M, Xie, G, Yeh, H.J.C, Liu, X, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 1998-08-06 | | Release date: | 1998-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the minor conformer of a DNA duplex containing a dG mismatch opposite a benzo[a]pyrene diol epoxide/dA adduct: glycosidic rotation from syn to anti at the modified deoxyadenosine.
Biochemistry, 36, 1997
|
|
1C2Q
 
 | |
3IDQ
 
 | | Crystal structure of S. cerevisiae Get3 at 3.7 Angstrom resolution | | Descriptor: | ATPase GET3, NICKEL (II) ION, ZINC ION | | Authors: | Suloway, C.J.M, Chartron, J.W, Zaslaver, M, Clemons Jr, W.M. | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4KS2
 
 | | Influenza Neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-carbamimidamido-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-carbamimidamido-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
