1S6H
 
 | | PORCINE TRYPSIN COMPLEXED WITH GUANIDINE-3-PROPANOL INHIBITOR | | Descriptor: | 4-(HYDROXYMETHYL)BENZAMIDINE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-23 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1SDV
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | Descriptor: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | Authors: | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2004-02-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
3I4J
 
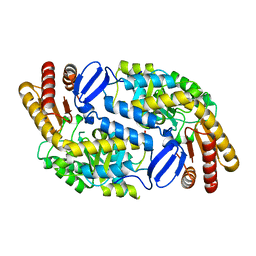 | | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans | | Descriptor: | Aminotransferase, class III, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans
To be Published
|
|
1LF7
 
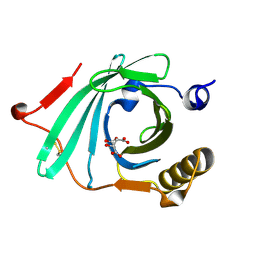 | | Crystal Structure of Human Complement Protein C8gamma at 1.2 A Resolution | | Descriptor: | CITRIC ACID, Complement Protein C8gamma | | Authors: | Ortlund, E, Parker, C.L, Schreck, S.F, Ginell, S, Minor, W, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-12 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of human complement protein C8gamma at 1.2 A resolution reveals a lipocalin fold and a distinct ligand binding site.
Biochemistry, 41, 2002
|
|
1SC4
 
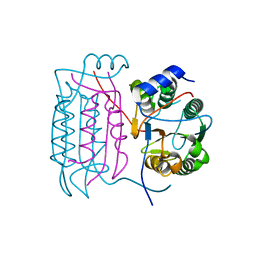 | |
3ZHP
 
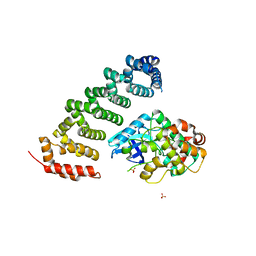 | | Human MST3 (STK24) in complex with MO25beta | | Descriptor: | CALCIUM-BINDING PROTEIN 39-LIKE, SERINE/THREONINE-PROTEIN KINASE 24, SULFATE ION | | Authors: | Elkins, J.M, Szklarz, M, Krojer, T, Mehellou, Y, Alessi, D.R, Chaikaud, A, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-12-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights Into the Activation of Mst3 by Mo25.
Biochem.Biophys.Res.Commun., 431, 2013
|
|
2GF5
 
 | | Structure of intact FADD (MORT1) | | Descriptor: | FADD protein | | Authors: | Carrington, P.E, Sandu, C, Wei, Y, Hill, J.M, Morisawa, G, Huang, T, Gavathiotis, E, Wei, Y, Werner, M.H. | | Deposit date: | 2006-03-21 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of FADD and Its Mode of Interaction with Procaspase-8
Mol.Cell, 22, 2006
|
|
3C8T
 
 | | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1 | | Descriptor: | Fumarate lyase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-13 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fumarate lyase from Mesorhizobium sp. BNC1.
To be Published
|
|
3PWF
 
 | | High resolution structure of the fully reduced form of rubrerythrin from P. furiosus | | Descriptor: | FE (II) ION, Rubrerythrin | | Authors: | Dillard, B.D, Demick, J.M, Adams, M.W, Lanzilotta, W.N. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A cryo-crystallographic time course for peroxide reduction by rubrerythrin from Pyrococcus furiosus.
J.Biol.Inorg.Chem., 16, 2011
|
|
2F84
 
 | |
1SH0
 
 | | Crystal Structure of Norwalk Virus Polymerase (Triclinic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
3I4S
 
 | | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN blr8122 FROM Bradyrhizobium japonicum | | Descriptor: | GLYCEROL, HISTIDINE TRIAD PROTEIN | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN FROM Bradyrhizobium japonicum
To be Published
|
|
1L7Q
 
 | | Ser117Ala Mutant of Bacterial Cocaine Esterase cocE | | Descriptor: | BENZOIC ACID, cocaine esterase | | Authors: | Turner, J.M, Larsen, N.A, Basran, A, Barbas III, C.F, Bruce, N.C, Wilson, I.A, Lerner, R.A. | | Deposit date: | 2002-03-16 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Biochemical characterization and structural analysis of a highly proficient cocaine
esterase
Biochemistry, 41, 2002
|
|
3ZCS
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(1-naphthoyl) -N-beta-D-glucopyranosyl urea determined at 2.07 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3HM7
 
 | | Crystal structure of allantoinase from Bacillus halodurans C-125 | | Descriptor: | Allantoinase, ZINC ION | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Miller, S, Wasserman, S.R, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Allantoinase from Bacillus Halodurans
To be Published
|
|
1LIE
 
 | |
1ON6
 
 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminotransferase (EXTL2) in complex with UDPGlcNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
3D19
 
 | | Crystal structure of a conserved metalloprotein from Bacillus cereus | | Descriptor: | Conserved metalloprotein, FE (III) ION, MAGNESIUM ION | | Authors: | Bonanno, J.B, Patskovsky, Y, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a conserved metalloprotein from Bacillus cereus.
To be Published
|
|
2FJM
 
 | | The structure of phosphotyrosine phosphatase 1B in complex with compound 2 | | Descriptor: | (4-{(2S,4E)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-2-[4-(METHOXYCARBONYL)PHENYL]-5-PHENYLPENT-4-ENYL}PHENYL)(DIFLUORO)METHYLPHOSPHONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Asante-Appiah, E, Patel, S, Desponts, C, Taylor, J.M, Lau, C, Dufresne, C, Therien, M, Friesen, R, Becker, J.W, Leblanc, Y, Scapin, G. | | Deposit date: | 2006-01-03 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformation-assisted inhibition of protein-tyrosine phosphatase-1B elicits inhibitor selectivity over T-cell protein-tyrosine phosphatase.
J.Biol.Chem., 281, 2006
|
|
3D46
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and L-tartrate | | Descriptor: | L(+)-TARTARIC ACID, MAGNESIUM ION, Putative galactonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and L-tartrate.
To be Published
|
|
3HY7
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with Marimastat | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
3CF1
 
 | | Structure of P97/vcp in complex with ADP/ADP.alfx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Transitional endoplasmic reticulum ATPase | | Authors: | Davies, J.M, Delabarre, B, Brunger, A.T, Weis, W.I. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.
Structure, 16, 2008
|
|
1NL9
 
 | | Potent, Selective Protein Tyrosine Phosphatase 1B Inhibitor Compound 12 Using a Linked-Fragment Strategy | | Descriptor: | 2-{[4-(2-ACETYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-NAPHTHALEN-1-YL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Pei, Z, Xin, Z, Lubben, T, Trevillyan, J.M, Stashko, M.A, Ballaron, S.J, Liang, H, Huang, F, Hutchins, C.W, Fesik, S.W, Jirousek, M.R. | | Deposit date: | 2003-01-06 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Selective Protein Tyrosine
Phosphatase 1B Inhibitor Using a
Linked-Fragment Strategy
J.Am.Chem.Soc., 125, 2003
|
|
3ZCV
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(indol-2- carbonyl)-N-beta-D-glucopyranosyl urea determined at 1.8 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-[(1H-indol-2-ylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3I14
 
 | |
