2GMR
 
 | | Photosynthetic reaction center mutant from Rhodobacter sphaeroides with Asp L210 replaced with Asn | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stachnik, J.M, Hermes, S, Gerwert, K, Hofmann, E. | | Deposit date: | 2006-04-07 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Proton uptake in the reaction center mutant L210DN from Rhodobacter sphaeroides via protonated water molecules.
Biochemistry, 45, 2006
|
|
1S81
 
 | | PORCINE TRYPSIN WITH NO INHIBITOR BOUND | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, DeRose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
3DO9
 
 | | Crystal structure of protein ba1542 from bacillus anthracis str.ames | | Descriptor: | UPF0302 protein BA_1542/GBAA1542/BAS1430 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Iizuka, M, Maletic, M, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-03 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Protein Ba1542 from Bacillus Anthracis Str.Ames.
To be Published
|
|
3I4K
 
 | | Crystal structure of Muconate lactonizing enzyme from Corynebacterium glutamicum | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Muconate lactonizing enzyme from Corynebacterium glutamicum
To be Published
|
|
4AOT
 
 | | Crystal Structure of Human Serine Threonine Kinase-10 (LOK) Bound to GW830263A | | Descriptor: | 1-(4-{methyl[2-({4-[(methylsulfonyl)methyl]phenyl}amino)pyrimidin-4-yl]amino}phenyl)-3-{3-[(4-methylpiperazin-1-yl)carbonyl]phenyl}urea, CHLORIDE ION, Serine/threonine-protein kinase 10 | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, Canning, P, von Delft, F, Yue, W, Liu, Y, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of Human Serine Threonine Kinase-10 (Lok) Bound to Gw830263A
To be Published
|
|
1SFP
 
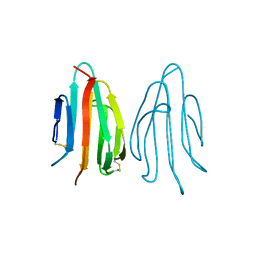 | | CRYSTAL STRUCTURE OF ACIDIC SEMINAL FLUID PROTEIN (ASFP) AT 1.9 A RESOLUTION: A BOVINE POLYPEPTIDE FROM THE SPERMADHESIN FAMILY | | Descriptor: | ASFP | | Authors: | Romao, M.J, Kolln, I, Dias, J.M, Carvalho, A.L, Romero, A, Varela, P.F, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
3P4W
 
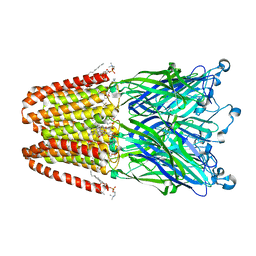 | | Structure of desflurane bound to a pentameric ligand-gated ion channel, GLIC | | Descriptor: | (2S)-2-(difluoromethoxy)-1,1,1,2-tetrafluoroethane, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nury, H, Van Renterghem, C, Weng, Y, Tran, A, Baaden, M, Dufresne, V, Changeux, J.P, Sonner, J.M, Delarue, M, Corringer, P.J. | | Deposit date: | 2010-10-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel
Nature, 469, 2011
|
|
3CR3
 
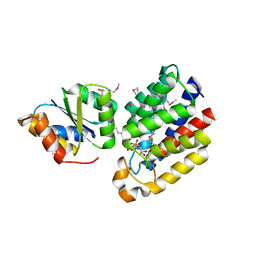 | | Structure of a transient complex between Dha-kinase subunits DhaM and DhaL from Lactococcus lactis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PTS-dependent dihydroxyacetone kinase, ... | | Authors: | Jeckelmann, J.M, Zurbriggen, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2008-04-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Three Lactococcus lactis Dihydroxyacetone Kinase Subunits and of a Transient Intersubunit Complex.
J.Biol.Chem., 283, 2008
|
|
1SL3
 
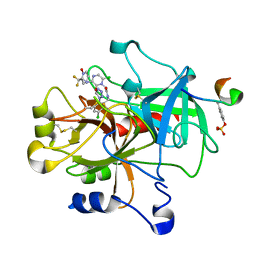 | | crystal structue of Thrombin in complex with a potent P1 heterocycle-Aryl based inhibitor | | Descriptor: | (2-[6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDOPYRIDIN-2-YL)ETHYL]AMINO}-2-OXOPYRAZIN-1(2H)-YL]-N-[5-CHLORO-2-(1H-TETRAZOL-1-YL)BENZYL]ACETAMIDE, Hirudin, thrombin | | Authors: | Young, M.B, Barrow, J.C, Glass, K.L, Lundell, G.F, Newton, C.L, Pellicore, J.M, Rittle, K.E, Selnick, H.G, Stauffer, K.J, Vacca, J.P, Williams, P.D, Bohn, D, Clayton, F.C, Cook, J.J, Krueger, J.A, Kuo, L.C, Lewis, S.D, Lucas, B.J, McMasters, D.R, Miller-Stein, C, Pietrak, B.L. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery and evaluation of potent P1 aryl heterocycle-based thrombin inhibitors
J.Med.Chem., 47, 2004
|
|
1SIH
 
 | | AGAO in covalent complex with the inhibitor MOBA ("4-(4-methylphenoxy)-2-butyn-1-amine") | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Guss, J.M, Langley, D.B, Duff, A.P. | | Deposit date: | 2004-02-29 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Differential Inhibition of Six Copper Amine Oxidases by a Family of 4-(Aryloxy)-2-butynamines: Evidence for a New Mode of Inactivation.
Biochemistry, 43, 2004
|
|
3GX0
 
 | | Crystal Structure of GSH-dependent Disulfide bond Oxidoreductase | | Descriptor: | GST-like protein yfcG, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Ladner, J.E, Harp, J.M, Wadington, M.C, Armstrong, R.N. | | Deposit date: | 2009-04-01 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase.
Biochemistry, 48, 2009
|
|
2HEP
 
 | | Solution NMR structure of the UPF0291 protein ynzC from Bacillus subtilis. Northeast Structural Genomics target SR384. | | Descriptor: | UPF0291 protein ynzC | | Authors: | Aramini, J.M, Swapna, G.V.T, Ho, C.K, Shetty, K, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-21 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SOS response protein YnzC from Bacillus subtilis
Proteins, 72, 2008
|
|
3CTA
 
 | | Crystal structure of riboflavin kinase from Thermoplasma acidophilum | | Descriptor: | Riboflavin kinase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of riboflavin kinase from Thermoplasma acidophilum.
To be Published
|
|
3CTP
 
 | | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with D-xylulofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, SODIUM ION, beta-D-xylulofuranose | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of periplasmic binding protein/LacI transcriptional regulator from Alkaliphilus metalliredigens QYMF complexed with L-xylulose.
To be Published
|
|
2GU8
 
 | |
1SQR
 
 | | Solution Structure of the 50S Ribosomal Protein L35AE from Pyrococcus furiosus. Northeast Structural Genomics Consortium Target PfR48. | | Descriptor: | 50S ribosomal protein L35Ae | | Authors: | Snyder, D.A, Aramini, J.M, Huang, Y.J, Xiao, R, Cort, J.R, Shastry, R, Ma, L.C, Liu, J, Rost, B, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 50S Ribosomal Protein L35AE from Pyrococcus furiosus: Northeast Structural Genomics Consortium Target PfR48
To be Published
|
|
1SPP
 
 | | THE CRYSTAL STRUCTURES OF TWO MEMBERS OF THE SPERMADHESIN FAMILY REVEAL THE FOLDING OF THE CUB DOMAIN | | Descriptor: | MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-I, MAJOR SEMINAL PLASMA GLYCOPROTEIN PSP-II | | Authors: | Romero, A, Romao, M.J, Varela, P.F, Kolln, I, Dias, J.M, Carvalho, A.L, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-19 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
3PDW
 
 | | Crystal structure of putative p-nitrophenyl phosphatase from Bacillus subtilis | | Descriptor: | ACETIC ACID, GLYCEROL, Uncharacterized hydrolase yutF | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Crystal structure of putative p-nitrophenyl phosphatase from Bacillus subtilis
To be Published
|
|
3PDY
 
 | |
2H48
 
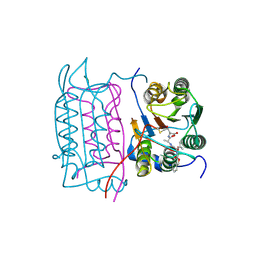 | | Crystal structure of human caspase-1 (Cys362->Ala, Cys364->Ala, Cys397->Ala) in complex with 3-[2-(2-benzyloxycarbonylamino-3-methyl-butyrylamino)-propionylamino]-4-oxo-pentanoic acid (z-VAD-FMK) | | Descriptor: | Caspase 1, isoform gamma, N-[(benzyloxy)carbonyl]-L-valyl-N-[(2S)-1-carboxy-4-fluoro-3-oxobutan-2-yl]-L-alaninamide | | Authors: | Scheer, J.M, Wells, J.A, Romanowski, M.J. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A common allosteric site and mechanism in caspases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1SC3
 
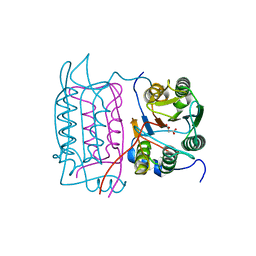 | | Crystal structure of the human caspase-1 C285A mutant in complex with malonate | | Descriptor: | Interleukin-1 beta convertase, MALONATE ION | | Authors: | Romanowski, M.J, Scheer, J.M, O'Brien, T, McDowell, R.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a ligand-free and malonate-bound human caspase-1: implications for the mechanism of substrate binding.
Structure, 12, 2004
|
|
3PC6
 
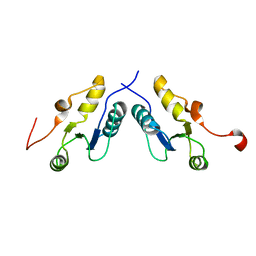 | |
2H54
 
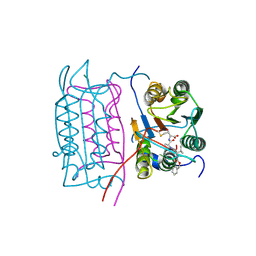 | |
3PC7
 
 | |
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
