1XJ7
 
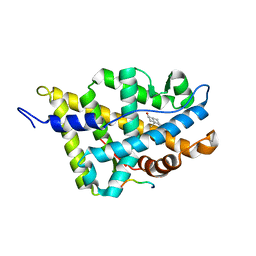 | | Complex Androgen Receptor LBD and RAC3 peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, RAC3 derived peptide | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Buehrer, B.M, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-09-22 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2XU1
 
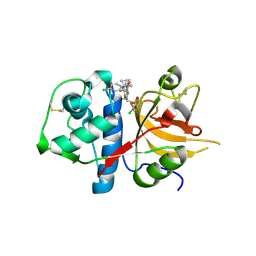 | | CATHEPSIN L WITH A NITRILE INHIBITOR | | Descriptor: | (2S,4R)-1-[1-(4-chlorophenyl)cyclopropyl]carbonyl-4-(2-chlorophenyl)sulfonyl-N-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, CATHEPSIN L1 | | Authors: | Banner, D.W, Benz, J.M, Steinbacher, S, Haap, W. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Investigation of Halogen Bonding in Protein-Ligand Interactions.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
4PXT
 
 | |
5J7T
 
 | |
1XXR
 
 | | Structure of a mannose-specific jacalin-related lectin from Morus Nigra in complex with mannose | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Rabijns, A, Barre, A, Van Damme, E.J.M, Peumans, W.J, De Ranter, C.J, Rouge, P. | | Deposit date: | 2004-11-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the jacalin-related lectin MornigaM from the black mulberry (Morus nigra) in complex with mannose
Febs J., 272, 2005
|
|
5J1B
 
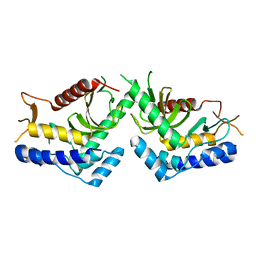 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - WT. | | Descriptor: | Cell wall assembly regulator SMI1 | | Authors: | Maveyraud, L, Batista, M, Martin-yken, H, Francois, J.M, Zerbib, D, Mourey, L. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biophysical characterization of Knr4, an intrinsically disordered hub protein form Saccharomyces cerevisiae
To Be Published
|
|
4PPV
 
 | |
4PED
 
 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
4PQH
 
 | | Crystal structure of glutathione transferase lambda1 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, SODIUM ION, glutathione transferase lambda1 | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
1ONY
 
 | | Oxalyl-Aryl-Amino Benzoic Acid inhibitors of PTP1B, compound 17 | | Descriptor: | 2-{[2-(2-CARBAMOYL-VINYL)-4-(2-METHANESULFONYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-PHENYL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
4PXU
 
 | | Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bipolar kinesin KRP-130 | | Authors: | Scholey, J.E, Nithianantham, S, Scholey, J.M, Al-Bassam, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers.
Elife, 3, 2014
|
|
3PI5
 
 | | Crystal Structure of Human Beta Secretase in Complex with BFG356 | | Descriptor: | (3S,4S,5R)-3-(3-bromo-4-hydroxybenzyl)-5-[(3-cyclopropylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1 | | Authors: | Rondeau, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based design, synthesis and SAR of cyclic hydroxyethylamine (HEA) BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1NML
 
 | | Di-haemic Cytochrome c Peroxidase from Pseudomonas nautica 617, form IN (pH 4.0) | | Descriptor: | CITRIC ACID, HEME C, di-haem cytochrome c peroxidase | | Authors: | Dias, J.M, Bonifacio, C, Alves, T, Pereira, A.S, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617
Structure, 12, 2004
|
|
4MN4
 
 | | Structural Basis for the MukB-topoisomerase IV Interaction | | Descriptor: | Chromosome partition protein MukB, DNA topoisomerase 4 subunit A | | Authors: | Vos, S.M, Stewart, N.K, Oakley, M.G, Berger, J.M. | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo.
Embo J., 32, 2013
|
|
4N48
 
 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with capped RNA fragment | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
4N6A
 
 | | Soybean Serine Acetyltransferase Apoenzyme | | Descriptor: | PHOSPHATE ION, Serine Acetyltransferase Apoenzyme | | Authors: | Yi, H, Dey, S, Kumaran, S, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone.
J.Biol.Chem., 288, 2013
|
|
4OIA
 
 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group P4322 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | Deposit date: | 2014-01-19 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3IWZ
 
 | | The c-di-GMP Responsive Global Regulator CLP Links Cell-Cell Signaling to Virulence Gene Expression in Xanthomonas campestris | | Descriptor: | Catabolite activation-like protein | | Authors: | Chin, K.H, Tu, Z.L, Tseng, Y.H, Dow, J.M, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2009-09-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris.
J.Mol.Biol., 396, 2010
|
|
4P0D
 
 | | The T6 backbone pilin of serotype M6 Streptococcus pyogenes has a modular three-domain structure decorated with variable loops and extensions | | Descriptor: | CALCIUM ION, IODIDE ION, Trypsin-resistant surface T6 protein | | Authors: | Young, P.G, Moreland, N.J, Loh, J.M, Bell, A, Atatoa-Carr, P, Proft, T, Baker, E.N. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Conservation, Variability, and Immunogenicity of the T6 Backbone Pilin of Serotype M6 Streptococcus pyogenes.
Infect.Immun., 82, 2014
|
|
1P6A
 
 | | STRUCTURAL BASIS FOR VARIATION IN ADENOVIRUS AFFINITY FOR THE CELLULAR RECEPTOR CAR (S489Y MUTANT) | | Descriptor: | Coxsackievirus and adenovirus receptor, Fiber protein | | Authors: | Howitt, J, Bewley, M.C, Graziano, V, Flanagan, J.M, Freimuth, P. | | Deposit date: | 2003-04-29 | | Release date: | 2004-05-11 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor.
J.Biol.Chem., 278, 2003
|
|
2L44
 
 | | C-terminal zinc knuckle of the HIVNCp7 | | Descriptor: | C-terminal Zinc Knucle of the HIV-NCP7, ZINC ION | | Authors: | Quintal, S.M.O, Viegas, A.J.M, Cabrita, E.J, Farrell, N.P, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
4HKQ
 
 | | XMRV reverse transcriptase in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*TP*GP*GP*AP*AP*TP*CP*A*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*TP*CP*TP*G)-3'), RNA (5'-R(*AP*AP*CP*AP*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*CP*AP*U)-3'), Reverse transcriptase/ribonuclease H p80 | | Authors: | Nowak, E, Potrzebowski, W, Konarev, P.V, Rausch, J.W, Bona, M.K, Svergun, D.I, Bujnicki, J.M, Le Grice, S.F.J, Nowotny, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid
Nucleic Acids Res., 41, 2013
|
|
3TMA
 
 | | Crystal structure of TrmN from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, methyltransferase | | Authors: | Fislage, M, Roovers, M, Tuszynska, I, Bujnicki, J.M, Droogmans, L, Versees, W. | | Deposit date: | 2011-08-31 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.
Nucleic Acids Res., 40, 2012
|
|
4HPU
 
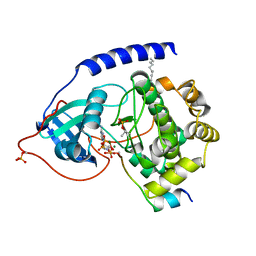 | | Crystal structure of the catalytic subunit of cAMP-dependent protein kinase displaying partial phosphoryl transfer of AMP-PNP onto a substrate peptide | | Descriptor: | MAGNESIUM ION, MYRISTIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Bastidas, A.C, Steichen, J.M, Wu, J, Taylor, S.S. | | Deposit date: | 2012-10-24 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phosphoryl transfer by protein kinase a is captured in a crystal lattice.
J.Am.Chem.Soc., 135, 2013
|
|
4EEU
 
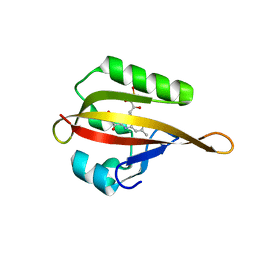 | | Crystal structure of phiLOV2.1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4068 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
