7RGM
 
 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH ATP, IMP, NAD+, OCTAMER-CENTERED | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, ... | | 著者 | Burrell, A.L, Kollman, J.M. | | 登録日 | 2021-07-15 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RGQ
 
 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH GTP, ATP, IMP, NAD+; INTERFACE-CENTERED | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | 著者 | Burrell, A.L, Kollman, J.M. | | 登録日 | 2021-07-15 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RFF
 
 | |
7RER
 
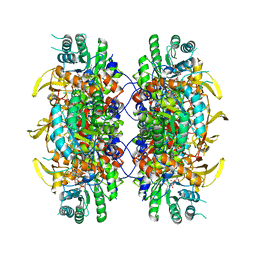 | | HUMAN IMPDH1 TREATED WITH ATP, IMP, AND NAD+ | | 分子名称: | INOSINIC ACID, Isoform 5 of Inosine-5'-monophosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Burrell, A.L, Kollman, J.M. | | 登録日 | 2021-07-13 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6VJL
 
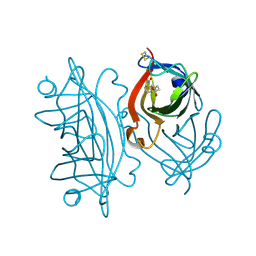 | | Streptavidin mutant M112 (G26C/A46C) | | 分子名称: | BIOTIN, Streptavidin | | 著者 | Marangoni, J.M, Wu, S.C, Fogen, D, Wong, S.L, Ng, K.K.S. | | 登録日 | 2020-01-16 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Engineering a disulfide-gated switch in streptavidin enables reversible binding without sacrificing binding affinity.
Sci Rep, 10, 2020
|
|
7RGI
 
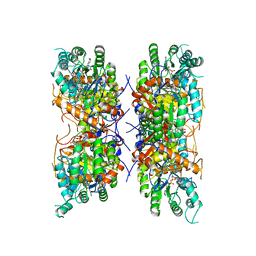 | | HUMAN RETINAL VARIANT IMPDH1(546) TREATED WITH GTP, ATP, IMP, NAD+; INTERFACE-CENTERED | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Burrell, A.L, Kollman, J.M. | | 登録日 | 2021-07-15 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RGD
 
 | | HUMAN RETINAL VARIANT IMPDH1(595) TREATED WITH GTP, ATP, IMP, NAD+, OCTAMER-CENTERED | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | 著者 | Burrell, A.L, Kollman, J.M. | | 登録日 | 2021-07-15 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | IMPDH1 retinal variants control filament architecture to tune allosteric regulation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6W1K
 
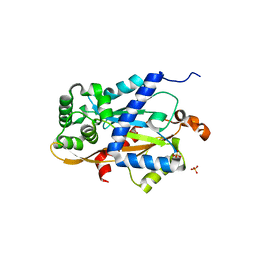 | | Crystal structure of the hydroxyglutarate synthase in complex with 2-oxoadipate from Oryza sativa | | 分子名称: | 2-OXOADIPIC ACID, Hydroxyglutarate synthase, NICKEL (II) ION, ... | | 著者 | Pereira, J.H, Thompson, M.G, Blake-Hedges, J.M, Keasling, J.D, Adams, P.D. | | 登録日 | 2020-03-04 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | An iron (II) dependent oxygenase performs the last missing step of plant lysine catabolism.
Nat Commun, 11, 2020
|
|
4ZWZ
 
 | | Engineered Carbonic Anhydrase IX mimic in complex with a glucosyl sulfamate inhibitor | | 分子名称: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ZINC ION, ... | | 著者 | Mahon, B.P, Lomelino, C.L, Driscoll, J.M, McKenna, R. | | 登録日 | 2015-05-19 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Mapping Selective Inhibition of the Cancer-Related Carbonic Anhydrase IX Using Structure-Activity Relationships of Glucosyl-Based Sulfamates.
J.Med.Chem., 58, 2015
|
|
6W1P
 
 | | RT XFEL structure of the one-flash state of Photosystem II (1F, S2-rich) at 2.26 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2020-03-04 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5A0G
 
 | | N-terminal thioester domain of surface protein from Clostridium perfringens | | 分子名称: | SURFACE ANCHORED PROTEIN | | 著者 | Walden, M, Edwards, J.M, Dziewulska, A.M, Kan, S.-Y, Schwarz-Linek, U, Banfield, M.J. | | 登録日 | 2015-04-20 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | An internal thioester in a pathogen surface protein mediates covalent host binding.
Elife, 4, 2015
|
|
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | 著者 | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | 登録日 | 2021-04-30 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.427 Å) | | 主引用文献 | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
6TW1
 
 | | Bat Influenza A polymerase termination complex with pyrophosphate using 44-mer vRNA template with mutated oligo(U) sequence | | 分子名称: | 5-oxidanyl-4-oxidanylidene-1-[(1-pyrrolo[2,3-b]pyridin-1-ylcyclopentyl)methyl]pyridine-3-carboxylic acid, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | 著者 | Wandzik, J.M, Kouba, T, Cusack, S. | | 登録日 | 2020-01-11 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 2020
|
|
7XLI
 
 | | Crystal structure of IsdB linker-NEAT2 bound to a nanobody (VHH) | | 分子名称: | CALCIUM ION, CHLORIDE ION, Iron-regulated surface determinant protein B, ... | | 著者 | Caaveiro, J.M.M, Valenciano-Bellido, S, Tsumoto, K. | | 登録日 | 2022-04-21 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Targeting hemoglobin receptors IsdH and IsdB of Staphylococcus aureus with a single VHH antibody inhibits bacterial growth.
J.Biol.Chem., 299, 2023
|
|
6U0W
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K133M at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | 登録日 | 2019-08-15 | | 公開日 | 2019-08-28 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2LXD
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for LMO2(LIM2)-Ldb1(LID) | | 分子名称: | Rhombotin-2,LIM domain-binding protein 1, ZINC ION | | 著者 | Dastmalchi, S, Wilkinson-White, L, Kwan, A.H, Gamsjaeger, R, Mackay, J.P, Matthews, J.M. | | 登録日 | 2012-08-20 | | 公開日 | 2012-09-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a tethered Lmo2(LIM2) /Ldb1(LID) complex.
Protein Sci., 21, 2012
|
|
6TWW
 
 | | Variant W229D/F290W-19 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) | | 分子名称: | ACETATE ION, Beta-Lactamase (GNCA4), FORMIC ACID, ... | | 著者 | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L. | | 登録日 | 2020-01-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
7L8I
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21) | | 分子名称: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | 著者 | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | 登録日 | 2020-12-31 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
7L8J
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21212) | | 分子名称: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | 著者 | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | 登録日 | 2020-12-31 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
7L8H
 
 | | EV68 3C protease (3Cpro) in Complex with Rupintrivir | | 分子名称: | 3C Protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | 著者 | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | 登録日 | 2020-12-31 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
6VKO
 
 | |
7XAR
 
 | | Crystal structure of 3C-like protease from SARS-CoV-2 in complex with covalent inhibitor | | 分子名称: | 3C-like proteinase, 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | 著者 | Caaveiro, J.M.M, Ochi, J, Takahashi, D, Ueda, T, Ojida, A. | | 登録日 | 2022-03-18 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease.
J.Med.Chem., 65, 2022
|
|
7XL0
 
 | | Crystal structure of Vobarilizumab at 1.70 Angstrom | | 分子名称: | GLYCEROL, Nanobody Vobarilizumab, SULFATE ION | | 著者 | Caaveiro, J.M.M, Mori, C, Kinoshita, S, Nakakido, M, Tsumoto, K. | | 登録日 | 2022-04-20 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular basis for thermal stability and affinity in a VHH: Contribution of the framework region and its influence in the conformation of the CDR3.
Protein Sci., 31, 2022
|
|
4Z4B
 
 | |
6W1V
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | 登録日 | 2020-03-04 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
