3JW3
 
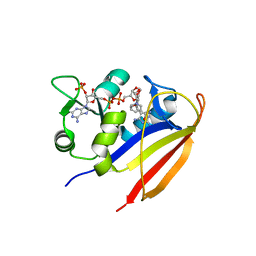 | |
3JWM
 
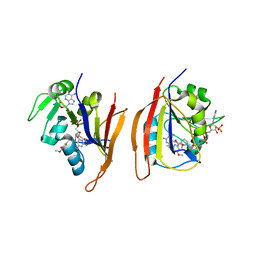 | | Crystal structure of Bacillus anthracis dihydrofolate reductase complexed with NADPH and (S)-2,4-diamino-5-(3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-methylpyrimidine (UCP114A) | | 分子名称: | 5-[(3S)-3-methoxy-3-(3,4,5-trimethoxyphenyl)prop-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | 登録日 | 2009-09-18 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Mutational Studies into Trimethoprim Resistance in Bacillus anthracis Dihydrofolate Reductase
To be Published
|
|
1SDU
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | 分子名称: | ACETATE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, SULFATE ION, ... | | 著者 | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2004-02-14 | | 公開日 | 2004-05-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
1SDT
 
 | | Crystal structures of HIV protease V82A and L90M mutants reveal changes in indinavir binding site. | | 分子名称: | CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, protease RETROPEPSIN | | 著者 | Mahalingam, B, Wang, Y.-F, Boross, P.I, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2004-02-14 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structures of HIV protease V82A and L90M
mutants reveal changes in the indinavir-binding site
Eur.J.Biochem., 271, 2004
|
|
3P8H
 
 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | 分子名称: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | 著者 | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-10-13 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
1SH3
 
 | | Crystal Structure of Norwalk Virus Polymerase (MgSO4 crystal form) | | 分子名称: | MAGNESIUM ION, RNA Polymerase | | 著者 | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | 登録日 | 2004-02-24 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
3C97
 
 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | 分子名称: | Signal transduction histidine kinase | | 著者 | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-02-15 | | 公開日 | 2008-03-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3OX4
 
 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 complexed with NAD cofactor | | 分子名称: | Alcohol dehydrogenase 2, FE (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | 登録日 | 2010-09-21 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
3H9M
 
 | | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii | | 分子名称: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, TRIETHYLENE GLYCOL, p-aminobenzoate synthetase, ... | | 著者 | Sampathkumar, P, Atwell, S, Wasserman, S, Do, J, Bain, K, Rutter, M, Gheyi, T, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-04-30 | | 公開日 | 2009-06-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Crystal structure of para-aminobenzoate synthetase, component I from Cytophaga hutchinsonii
To be Published
|
|
1SFK
 
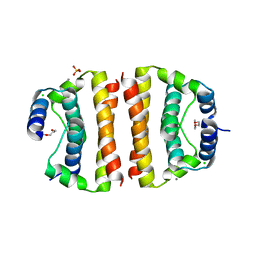 | | Core (C) protein from West Nile Virus, subtype Kunjin | | 分子名称: | CALCIUM ION, CHLORIDE ION, Core protein, ... | | 著者 | Dokland, T, Walsh, M, Mackenzie, J.M, Khromykh, A.A, Ee, K.-H, Wang, S. | | 登録日 | 2004-02-19 | | 公開日 | 2004-08-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | West nile virus core protein; tetramer structure and ribbon formation
Structure, 12, 2004
|
|
3KAL
 
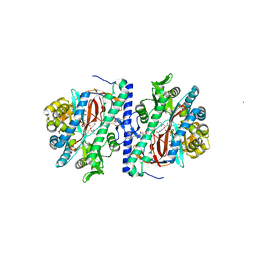 | | Structure of homoglutathione synthetase from Glycine max in closed conformation with homoglutathione, ADP, a sulfate ion, and three magnesium ions bound | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, D-gamma-glutamyl-L-cysteinyl-beta-alanine, MAGNESIUM ION, ... | | 著者 | Galant, A, Arkus, K.A.J, Zubieta, C, Cahoon, R.E, Jez, J.M. | | 登録日 | 2009-10-19 | | 公開日 | 2009-12-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Evolution of Product Diversity in Soybean Glutathione Biosynthesis.
Plant Cell, 21, 2009
|
|
3BQT
 
 | |
3J3I
 
 | | Penicillium chrysogenum virus (PcV) capsid structure | | 分子名称: | Capsid protein | | 著者 | Luque, D, Gomez-Blanco, J, Garriga, D, Brilot, A, Gonzalez, J.M, Havens, W.H, Carrascosa, J.L, Trus, B.L, Verdaguer, N, Grigorieff, N, Ghabrial, S.A, Caston, J.R. | | 登録日 | 2013-03-08 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3HCW
 
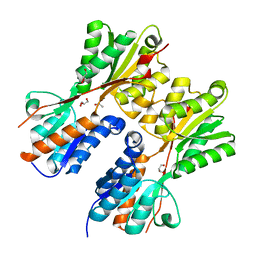 | | CRYSTAL STRUCTURE OF PROBABLE maltose operon transcriptional repressor malR FROM STAPHYLOCOCCUS AREUS | | 分子名称: | GLYCEROL, Maltose operon transcriptional repressor | | 著者 | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-05-06 | | 公開日 | 2009-05-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Maltose Operon Transcriptional Repressor from Staphylococcus Aureus
To be Published
|
|
1O24
 
 | | Crystal structure of Thymidylate Synthase Complementing Protein (TM0449) from Thermotoga maritima at 2.0 A resolution | | 分子名称: | Thymidylate synthase thyX | | 著者 | Mathews, I.I, Deacon, A.M, Canaves, J.M, McMullan, D, Lesley, S.A, Agarwalla, S, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2003-02-18 | | 公開日 | 2003-06-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional Analysis of Substrate and Cofactor Complex Structures of a Thymidylate Synthase-Complementing Protein
Structure, 11, 2003
|
|
3J9F
 
 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | 著者 | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | 登録日 | 2015-01-15 | | 公開日 | 2015-02-11 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3I4S
 
 | | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN blr8122 FROM Bradyrhizobium japonicum | | 分子名称: | GLYCEROL, HISTIDINE TRIAD PROTEIN | | 著者 | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-07-02 | | 公開日 | 2009-07-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN FROM Bradyrhizobium japonicum
To be Published
|
|
3CZB
 
 | | Crystal structure of putative transglycosylase from Caulobacter crescentus | | 分子名称: | Putative transglycosylase, SULFATE ION | | 著者 | Ramagopal, U.A, Chattopadhyay, K, Toro, R, Wasserman, S, Freeman, J, Logan, C, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-04-28 | | 公開日 | 2008-06-10 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of putative transglycosylase from Caulobacter crescentus.
To be Published
|
|
4A8O
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | 分子名称: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | 著者 | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | 登録日 | 2011-11-21 | | 公開日 | 2012-07-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
3HY7
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with Marimastat | | 分子名称: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | 著者 | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | 登録日 | 2009-06-22 | | 公開日 | 2009-07-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
3D2Z
 
 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the product L-Ala-D-gamma-Glu-L-Lys | | 分子名称: | CHLORIDE ION, L-Ala-D-gamma-Glu-L-Lys peptide, N-acetylmuramoyl-L-alanine amidase amiD, ... | | 著者 | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | 登録日 | 2008-05-09 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3Q6P
 
 | | Salivary protein from Lutzomyia longipalpis. Selenomethionine derivative | | 分子名称: | 43.2 kDa salivary protein, CITRIC ACID | | 著者 | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | 登録日 | 2011-01-03 | | 公開日 | 2011-07-27 | | 最終更新日 | 2019-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
1SH0
 
 | | Crystal Structure of Norwalk Virus Polymerase (Triclinic) | | 分子名称: | RNA Polymerase | | 著者 | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | 登録日 | 2004-02-24 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
3I14
 
 | |
3JVX
 
 | | Crystal structure of Bacillus anthracis dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-(3-(3,4,5-trimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120A) | | 分子名称: | 6-ethyl-5-[3-(3,4,5-trimethoxyphenyl)prop-1-yn-1-yl]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Anderson, A.C, Beierlein, J.M, Karri, N.G. | | 登録日 | 2009-09-17 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Mutational Studies into Trimethoprim Resistance in Bacillus anthracis Dihydrofolate Reductase
To be Published
|
|
