8QO5
 
 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | SARS-CoV-2-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QF5
 
 | |
8QF3
 
 | |
4LZD
 
 | | Human DNA polymerase mu- Apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4MDB
 
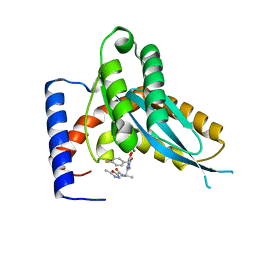 | | Structure of Mos1 transposase catalytic domain and Raltegravir with Mg | | Descriptor: | MAGNESIUM ION, Mariner Mos1 transposase, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide | | Authors: | Richardson, J.M. | | Deposit date: | 2013-08-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Mos1 Transposase Inhibition by the Anti-retroviral Drug Raltegravir.
Acs Chem.Biol., 9, 2014
|
|
8QF4
 
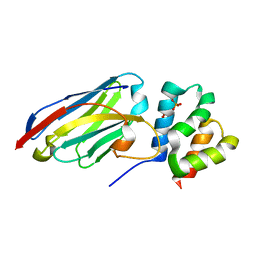 | |
8Q5P
 
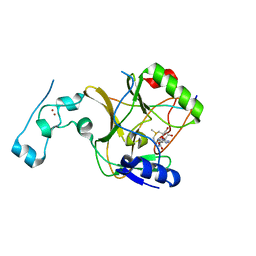 | | Structure of the lysine methyltransferase SETD2 in complex with a peptide derived from human tyrosine kinase ACK1 | | Descriptor: | Activated CDC42 kinase 1, Histone-lysine N-methyltransferase SETD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Mechaly, A, Le Coadou, L, Dupret, J.M, Haouz, A, Rodrigues Lima, F. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Structural and enzymatic evidence for the methylation of the ACK1 tyrosine kinase by the histone lysine methyltransferase SETD2.
Biochem.Biophys.Res.Commun., 695, 2024
|
|
5JYG
 
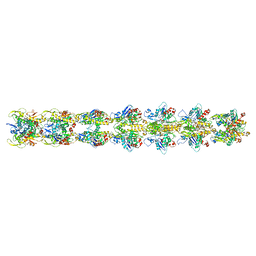 | |
4LYJ
 
 | | Crystal Structure of small molecule vinylsulfonamide 9 covalently bound to K-Ras G12C, alternative space group | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-31 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LZG
 
 | | Binary complex of human DNA Polymerase Mu with DNA | | Descriptor: | CHLORIDE ION, DNA-directed DNA/RNA polymerase mu, GLYCEROL, ... | | Authors: | Moon, A.F, Pryor, J.M, Ramsden, D.A, Kunkel, T.A, Bebenek, K, Pedersen, L.C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Sustained active site rigidity during synthesis by human DNA polymerase mu.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4MDA
 
 | | Structure of Mos1 transposase catalytic domain and Raltegravir with Mn | | Descriptor: | MANGANESE (II) ION, Mariner Mos1 transposase, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide | | Authors: | Richardson, J.M. | | Deposit date: | 2013-08-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Mos1 Transposase Inhibition by the Anti-retroviral Drug Raltegravir.
Acs Chem.Biol., 9, 2014
|
|
8Q6X
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
8Q6Y
 
 | | Crystal structure of Cytochrome P450 GymB5 from Streptomyces katrae in complex with cYY and Hypoxanthine | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, 1,2-ETHANEDIOL, Cytochrome P450, ... | | Authors: | Freytag, J, Mueller, J.M, Stehle, T, Zocher, G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Intramolecular Coupling and Nucleobase Transfer - How Cytochrome P450 Enzymes GymBx Establish Their Chemoselectivity
Chemcatchem, 16, 2024
|
|
4LV6
 
 | | Crystal Structure of small molecule disulfide 4 covalently bound to K-Ras G12C | | Descriptor: | 1-[(2,4-dichlorophenoxy)acetyl]-N-(2-sulfanylethyl)piperidine-4-carboxamide, CALCIUM ION, GTPase KRas, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-26 | | Release date: | 2013-11-27 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
5K8X
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor U3 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor U3
To Be Published
|
|
4M1N
 
 | | Crystal structure of Plasmodium falciparum ubiquitin conjugating enzyme UBC9 | | Descriptor: | SODIUM ION, Ubiquitin conjugating enzyme UBC9 | | Authors: | Boucher, L.E, Reiter, K.H, Bosch, J, Matunis, J.M. | | Deposit date: | 2013-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Biochemically Distinct Properties of the Small Ubiquitin-related Modifier (SUMO) Conjugation Pathway in Plasmodium falciparum.
J.Biol.Chem., 288, 2013
|
|
4M1U
 
 | | The crystal structure of Stx2 and a disaccharide ligand | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-methyl beta-D-galactopyranoside, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 A-subunit, ... | | Authors: | Yin, J, James, M.N.G, Jacobson, J.M, Kitov, P.I, Bundle, D.R, Mulvey, G, Armstrong, G. | | Deposit date: | 2013-08-04 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The crystal structure of shiga toxin type 2 with bound disaccharide guides the design of a heterobifunctional toxin inhibitor.
J.Biol.Chem., 289, 2014
|
|
8QBF
 
 | | Compact state - Pil1 dimer with lipid headgroups fitted in native eisosome lattice bound to plasma membrane microdomain | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOSERINE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8QB7
 
 | |
8QB8
 
 | |
5K8W
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor U2 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor U2
To Be Published
|
|
8QBB
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/-sterol (DOPC, DOPE, DOPS, PI(4,5)P2 50:20:20:10) | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
4M1O
 
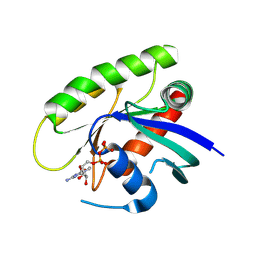 | | Crystal Structure of small molecule vinylsulfonamide 7 covalently bound to K-Ras G12C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras GTPase, N-(1-{[(5,7-dichloro-2,2-dimethyl-1,3-benzodioxol-4-yl)oxy]acetyl}piperidin-4-yl)ethanesulfonamide | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-08-03 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
8QBD
 
 | | Helical reconstruction of yeast eisosome protein Pil1 bound to membrane composed of lipid mixture +PIP2/+sterol (DOPC, DOPE, DOPS, cholesterol, PI(4,5)P2 35:20:20:15:10) | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sphingolipid long chain base-responsive protein PIL1 | | Authors: | Kefauver, J.M, Zou, L, Desfosses, A, Loewith, R.J. | | Deposit date: | 2023-08-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM architecture of a near-native stretch-sensitive membrane microdomain.
Nature, 632, 2024
|
|
8Q3W
 
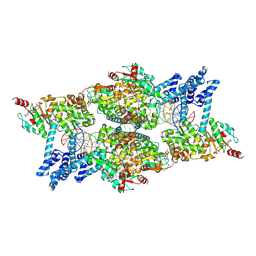 | | ATP-bound IstB in complex to duplex DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (48-MER) Target DNA FW, DNA (48-MER) Traget DNA Rv, ... | | Authors: | de la Gandara, A, Spinola-Amilibia, M, Araujo-Bazan, L, Nunez-Ramirez, R, Berger, J.M, Arias-Palomo, E. | | Deposit date: | 2023-08-04 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Molecular basis for transposase activation by a dedicated AAA+ ATPase.
Nature, 630, 2024
|
|
