2FCF
 
 | | The crystal structure of the 7th PDZ domain of MPDZ (MUPP-1) | | Descriptor: | Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Berridge, G, Johansson, C, Colebrook, S, Salah, E, Burgess, N, Smee, C, Savitsky, P, Bray, J, Schoch, G, Phillips, C, Gileadi, C, Soundarajan, M, Yang, X, Elkins, J.M, Gorrec, F, Turnbull, A, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
3CTD
 
 | | Crystal structure of a putative AAA family ATPase from Prochlorococcus marinus subsp. pastoris | | Descriptor: | Putative ATPase, AAA family | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative AAA family ATPase from Prochlorococcus marinus subsp. pastoris.
To be Published
|
|
4AU4
 
 | | Crystal Structure of Hsp47 | | Descriptor: | SERPIN PEPTIDASE INHIBITOR, CLADE H (HEAT SHOCK PROTEIN 47), MEMBER 1, ... | | Authors: | Widmer, C, Gebauer, J.M, Brunstein, E, Rodenbaum, S, Zaucke, F, Drogemuller, C, Leeb, T, Baumann, U. | | Deposit date: | 2012-05-14 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Molecular Basis for the Action of the Collagen-Specific Chaperone Hsp47/Serpinh1 and its Structure-Specific Client Recognition.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
1MQ3
 
 | | Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine Template Paired with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*(8OG)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | Authors: | Krahn, J.M, Beard, W.A, Miller, H, Grollman, A.P, Wilson, S.H. | | Deposit date: | 2002-09-13 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of DNA Polymerase beta with the Mutagenic DNA Lesion 8-oxodeoxyguanine Reveals
Structural Insights into its Coding Potential
Structure, 11, 2003
|
|
3GH1
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | PHOSPHATE ION, Predicted nucleotide-binding protein | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae.
To be Published
|
|
4B53
 
 | | Crystal structure of the isolated IgG4 CH3 domain | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Davies, A.M, Rispens, T, den Bleker, T.H, McDonnell, J.M, Gould, H.J, Aalberse, R.C, Sutton, B.J. | | Deposit date: | 2012-08-02 | | Release date: | 2012-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Human Igg4 C(H)3 Dimer Reveals the Role of Arg409 in the Mechanism of Fab-Arm Exchange.
Mol.Immunol., 54, 2012
|
|
1M13
 
 | | Crystal Structure of the Human Pregane X Receptor Ligand Binding Domain in Complex with Hyperforin, a Constituent of St. John's Wort | | Descriptor: | 4-HYDROXY-5-ISOBUTYRYL-6-METHYL-1,3,7-TRIS-(3-METHYL-BUT-2-ENYL)-6-(4-METHYL-PENT-3-ENYL)-BICYCLO[3.3.1]NON-3-ENE-2,9-DIONE, Orphan Nuclear Receptor PXR | | Authors: | Watkins, R.E, Maglich, J.M, Moore, L.B, Wisely, G.B, Noble, S.M, Davis-Searles, P.R, Lambert, M.H, Kliewer, S.A, Redinbo, M.R. | | Deposit date: | 2002-06-17 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 A Crystal Structure of Human PXR in Complex with the St. John's Wort Compound Hyperforin
Biochemistry, 42, 2003
|
|
2GJ5
 
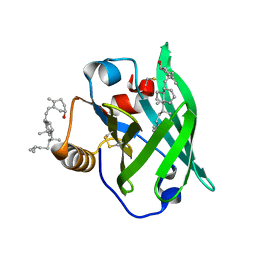 | | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Beta-lactoglobulin | | Authors: | Yang, M.C, Guan, H.H, Liu, M.Y, Yang, J.M, Chen, W.L, Chen, C.J, Mao, S.J. | | Deposit date: | 2006-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin.
Proteins, 71, 2008
|
|
3CKX
 
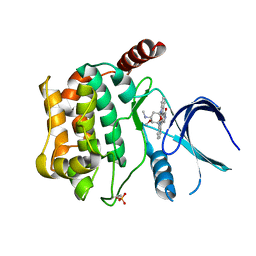 | | Crystal structure of sterile 20-like kinase 3 (MST3, STK24) in complex with staurosporine | | Descriptor: | STAUROSPORINE, Serine/threonine-protein kinase 24 | | Authors: | Antonysamy, S.S, Burley, S.K, Buchanan, S, Chau, F, Feil, I, Wu, L, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-17 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of sterile 20-like kinase 3 (MST3, STK24) in complex with staurosporine.
To be Published
|
|
2GL0
 
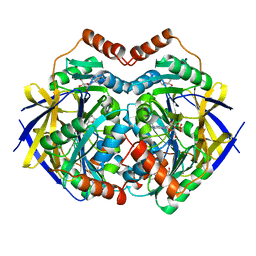 | | Structure of PAE2307 in complex with adenosine | | Descriptor: | ADENOSINE, PHOSPHATE ION, conserved hypothetical protein | | Authors: | Lott, J.S, Paget, B, Johnston, J.M, Baker, E.N. | | Deposit date: | 2006-04-04 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure of an Ancient Conserved Domain Establishes a Structural Basis for Stable Histidine Phosphorylation and Identifies a New Family of Adenosine-specific Kinases.
J.Biol.Chem., 281, 2006
|
|
1Q5H
 
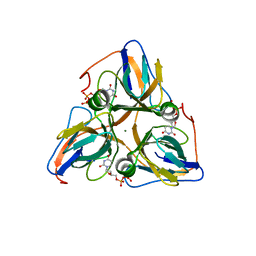 | | Human dUTP Pyrophosphatase complex with dUDP | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, dUTP pyrophosphatase | | Authors: | Mol, C.D, Harris, J.M, McIntosh, E.M, Tainer, J.A. | | Deposit date: | 2003-08-07 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human dUTP pyrophosphatase: uracil recognition by a Beta hairpin and active sites formed by three separate subunits
Structure, 4, 1996
|
|
1N5Y
 
 | | HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3', 5'-D(*AP*TP*GP*C*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, P, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 Reverse Transcriptase with Pre-Translocation and Post-Translocation AZTMP-Terminated DNA
Embo J., 21, 2002
|
|
4BIJ
 
 | | Threading model of T7 large terminase | | Descriptor: | DNA MATURASE B | | Authors: | Dauden, M.I, Martin-Benito, J, Sanchez-Ferrero, J.C, Pulido-Cid, M, Valpuesta, J.M, Carrascosa, J.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Large Terminase Conformational Change Induced by Connector Binding in Bacteriophage T7
J.Biol.Chem., 288, 2013
|
|
3G79
 
 | | Crystal structure of NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase from Methanosarcina mazei Go1 | | Descriptor: | NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of NDP-N-acetyl-D-galactosaminuronic acid dehydrogenase from Methanosarcina mazei Go1
To be Published
|
|
3CS3
 
 | | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis | | Descriptor: | GLYCEROL, SULFATE ION, Sugar-binding transcriptional regulator, ... | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis.
To be Published
|
|
3CT4
 
 | | Structure of Dha-kinase subunit DhaK from L. Lactis | | Descriptor: | Dihydroxyacetone, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Jeckelmann, J.M, Zurbriggen, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2008-04-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | X-ray Structures of the Three Lactococcus lactis Dihydroxyacetone Kinase Subunits and of a Transient Intersubunit Complex.
J.Biol.Chem., 283, 2008
|
|
2GVM
 
 | | Crystal structure of hydrophobin HFBI with detergent | | Descriptor: | Hydrophobin-1, LAURYL DIMETHYLAMINE-N-OXIDE, ZINC ION | | Authors: | Hakanpaa, J.M, Rouvinen, J. | | Deposit date: | 2006-05-03 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two crystal structures of Trichoderma reesei hydrophobin HFBI--The structure of a protein amphiphile with and without detergent interaction.
Protein Sci., 15, 2006
|
|
1RHL
 
 | | RIBONUCLEASE T1 COMPLEXED WITH 2'GMP/G23A MUTANT | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Huyghues-Despointes, B.M.P, Langhorst, U, Steyaert, J, Pace, C.N, Scholtz, J.M. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hydrogen-exchange stabilities of RNase T1 and variants with buried and solvent-exposed Ala --> Gly mutations in the helix.
Biochemistry, 38, 1999
|
|
3GG7
 
 | | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, ... | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans
To be Published
|
|
3Q2B
 
 | | Crystal Structure of an Actin Depolymerizing Factor | | Descriptor: | BETA-MERCAPTOETHANOL, Cofilin/actin-depolymerizing factor homolog 1, D(-)-TARTARIC ACID | | Authors: | Wong, W, Clarke, O.B, Gulbis, J.M, Baum, J. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Minimal requirements for actin filament disassembly revealed by structural analysis of malaria parasite actin-depolymerizing factor 1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1N7I
 
 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and the inhibitor LY134046 | | Descriptor: | 8,9-DICHLORO-2,3,4,5-TETRAHYDRO-1H-BENZO[C]AZEPINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
3G5L
 
 | | Crystal structure of putative S-adenosylmethionine dependent methyltransferase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Putative S-adenosylmethionine dependent methyltransferase | | Authors: | Patskovsky, Y, Sampathkumar, P, Gilmore, M, Miller, S, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of S-Adenosylmethionine Dependent Methyltransferase from Listeria Monocytogenes
To be Published
|
|
2H4W
 
 | |
2H6Y
 
 | |
