4FUN
 
 | |
4GP0
 
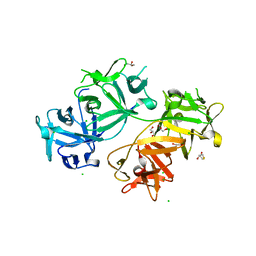 | | The crystal structure of human fascin 1 R149A K150A R151A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GOV
 
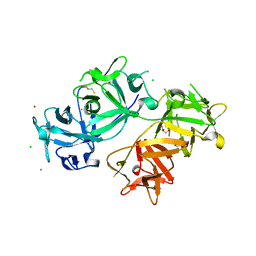 | | The crystal structure of human fascin 1 S39D mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
7CIX
 
 | |
7CIU
 
 | |
7CIW
 
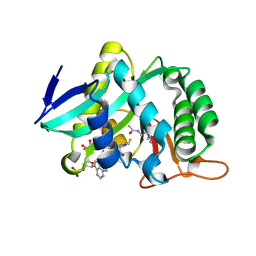 | |
6AC7
 
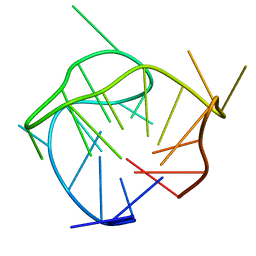 | | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*TP*CP*CP*GP*AP*GP*GP*CP*GP*GP*GP*GP*CP*TP*TP*GP*GP*G)-3' | | Authors: | Sengar, A, Vandana, J.J, Chambers, V.S, Di Antonio, M, Winnerdy, F.R, Balasubramanian, S, Phan, A.T. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter.
Nucleic Acids Res., 47, 2019
|
|
7CIV
 
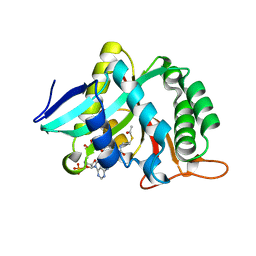 | |
5WAH
 
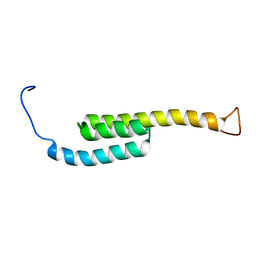 | | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN | | Descriptor: | IgA FC receptor | | Authors: | ELETSKY, A, CHEN, C, FONG, J.J, NIZET, V, VARKI, A, PRESTEGARD, J.H. | | Deposit date: | 2017-06-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF SIGLEC-5 BINDING DOMAIN FROM STREPTOCOCCAL BETA PROTEIN
To Be Published
|
|
5YFP
 
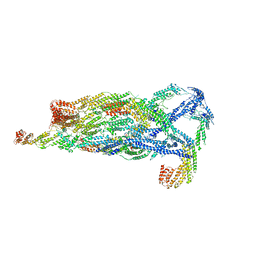 | | Cryo-EM Structure of the Exocyst Complex | | Descriptor: | Exocyst complex component EXO70, Exocyst complex component EXO84, Exocyst complex component SEC10, ... | | Authors: | Mei, K, Li, Y, Wang, S, Shao, G, Wang, J, Ding, Y, Luo, G, Yue, P, Liu, J.J, Wang, X, Dong, M.Q, Guo, W, Wang, H.W. | | Deposit date: | 2017-09-21 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the exocyst complex
Nat. Struct. Mol. Biol., 25, 2018
|
|
5Y66
 
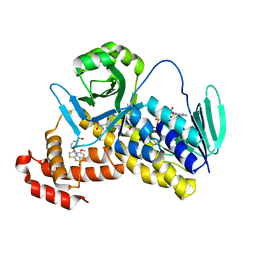 | | Crystal structure of Pseudomonas fluorescens Kynurenine 3-monooxygenase in complex with L-KYN and Ro61-8048 | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xiang, Y, Gao, J.J, Zhu, D.Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Biochemistry and structural studies of kynurenine 3-monooxygenase reveal allosteric inhibition by Ro 61-8048
FASEB J., 32, 2018
|
|
5XBK
 
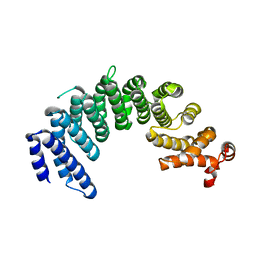 | | Crystal structure of human Importin4 | | Descriptor: | Importin-4, histone H3 | | Authors: | Song, J.J, Yoon, J. | | Deposit date: | 2017-03-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.223 Å) | | Cite: | Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding
J. Mol. Biol., 430, 2018
|
|
2OOM
 
 | | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA/RNA aptamer | | Descriptor: | RNA 16-mer, TAR RNA element of HIV-1 | | Authors: | Lebars, I, Richard, T, Di Primo, C, Toulme, J.J. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer
Nucleic Acids Res., 35, 2007
|
|
4FUO
 
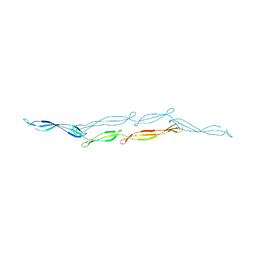 | |
4HPK
 
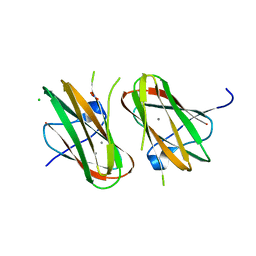 | | Crystal structure of Clostridium histolyticum colg collagenase collagen-binding domain 3B at 1.35 Angstrom resolution in presence of calcium nitrate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagenase, ... | | Authors: | Philominathan, S.T.L, Wilson, J.J, Bauer, R, Matsushita, O, Sakon, J. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
4HIP
 
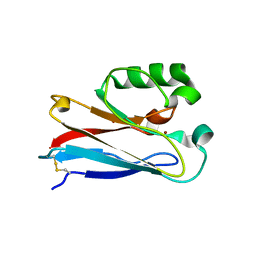 | |
4GDE
 
 | |
4GGK
 
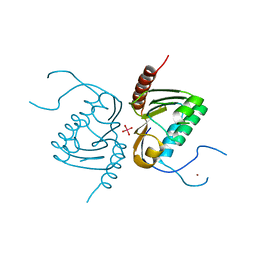 | | Crystal structure of Zucchini from mouse (mZuc / PLD6 / MitoPLD) bound to tungstate | | Descriptor: | Mitochondrial cardiolipin hydrolase, TUNGSTATE(VI)ION, ZINC ION | | Authors: | Ipsaro, J.J, Haase, A.D, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis.
Nature, 491, 2012
|
|
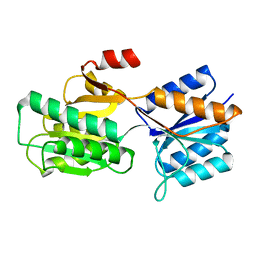
6ABP
 
 | |
4FEL
 
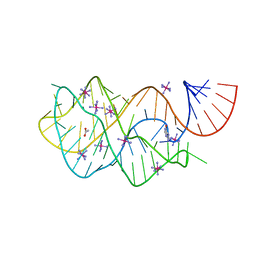 | | Crystal structure of the U25A/A46G mutant of the xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), HYPOXANTHINE, ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
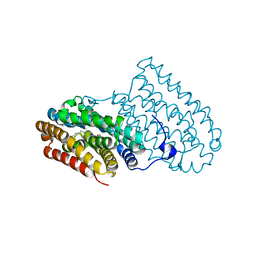
4HR4
 
 | |
4HR5
 
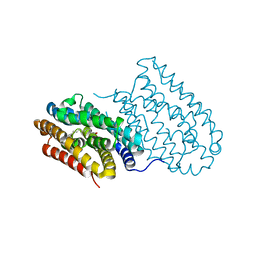 | | R2-like ligand-binding oxidase without metal cofactor | | Descriptor: | PALMITIC ACID, Ribonuleotide reductase small subunit | | Authors: | Griese, J.J, Hogbom, M. | | Deposit date: | 2012-10-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7CRB
 
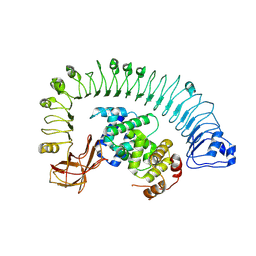 | | Cryo-EM structure of plant NLR RPP1 LRR-ID domain in complex with ATR1 | | Descriptor: | Avirulence protein ATR1, NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
2HYN
 
 | | Complete ensemble of NMR structures of unphosphorylated human phospholamban pentamer | | Descriptor: | Cardiac phospholamban | | Authors: | Potluri, S, Yan, A.K, Chou, J.J, Donald, B.R, Bailey-Kellogg, C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure determination of symmetric homo-oligomers by a complete search of symmetry configuration space, using NMR restraints and van der Waals packing.
Proteins, 65, 2006
|
|
4HHG
 
 | | Crystal structure of the Pseudomonas aeruginosa azurin, RuH107NO YOH109 | | Descriptor: | Azurin, COPPER (II) ION, DELTA-BIS(2,2'-BIPYRIDINE)IMIDAZOLE RUTHENIUM (II) | | Authors: | Herrera, N, Warren, J.J, Gray, H.B. | | Deposit date: | 2012-10-09 | | Release date: | 2012-11-21 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Electron Flow through Nitrotyrosinate in Pseudomonas aeruginosa Azurin.
J.Am.Chem.Soc., 135, 2013
|
|
